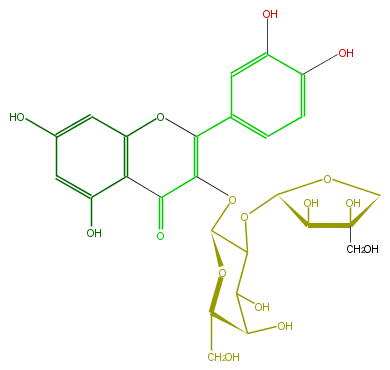
Mol:FL5FACGA0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0946 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0946 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0946 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0946 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3936 -0.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3936 -0.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6926 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6926 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6926 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6926 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3936 1.3633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3936 1.3633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9915 -0.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9915 -0.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2905 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2905 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2905 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2905 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9915 1.3633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9915 1.3633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9915 -1.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9915 -1.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4102 1.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4102 1.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1247 0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1247 0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8392 1.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8392 1.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8392 2.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8392 2.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1247 2.6006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1247 2.6006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4102 2.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4102 2.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7954 1.3632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7954 1.3632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4400 -0.2880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4400 -0.2880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3936 -0.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3936 -0.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6381 2.6494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6381 2.6494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7397 -1.3576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7397 -1.3576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0053 -0.9336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0053 -0.9336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2383 -1.7489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2383 -1.7489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0053 -2.5693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0053 -2.5693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7397 -2.9934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7397 -2.9934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5067 -2.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5067 -2.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6875 -0.6387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6875 -0.6387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9681 -2.4444 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9681 -2.4444 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4238 -2.8101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4238 -2.8101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3269 -0.2055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3269 -0.2055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9457 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9457 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7913 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7913 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3894 -0.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3894 -0.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3375 0.1245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3375 0.1245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9457 -0.3481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9457 -0.3481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7913 -0.3468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7913 -0.3468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1247 3.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1247 3.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0053 -3.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0053 -3.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7299 -2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7299 -2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7913 -1.2785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7913 -1.2785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7954 -1.5483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7954 -1.5483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 16 38 1 0 0 0 0 | + | 16 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 25 39 1 0 0 0 0 | + | 25 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 0.0000 0.8561 | + | M SBV 1 44 0.0000 0.8561 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 0.0000 0.4542 | + | M SBV 2 46 0.0000 0.4542 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGA0002 | + | ID FL5FACGA0002 |
| − | FORMULA C26H28O16 | + | FORMULA C26H28O16 |
| − | EXACTMASS 596.137734848 | + | EXACTMASS 596.137734848 |
| − | AVERAGEMASS 596.49092 | + | AVERAGEMASS 596.49092 |
| − | SMILES c(c5O)c(c(c(c5)3)C(C(=C(c(c4)ccc(O)c4O)O3)OC(O2)C(C(O)C(C(CO)2)O)OC(C(O)1)OCC1(O)CO)=O)O | + | SMILES c(c5O)c(c(c(c5)3)C(C(=C(c(c4)ccc(O)c4O)O3)OC(O2)C(C(O)C(C(CO)2)O)OC(C(O)1)OCC1(O)CO)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-3.0946 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0946 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3936 -0.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6926 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6926 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3936 1.3633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9915 -0.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2905 0.1491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2905 0.9585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9915 1.3633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9915 -1.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4102 1.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1247 0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8392 1.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8392 2.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1247 2.6006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4102 2.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7954 1.3632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4400 -0.2880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3936 -0.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6381 2.6494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7397 -1.3576 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0053 -0.9336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2383 -1.7489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0053 -2.5693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7397 -2.9934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5067 -2.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6875 -0.6387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9681 -2.4444 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4238 -2.8101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3269 -0.2055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9457 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7913 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3894 -0.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3375 0.1245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9457 -0.3481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7913 -0.3468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1247 3.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0053 -3.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7299 -2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7913 -1.2785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7954 -1.5483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 31 1 0 0 0 0
32 36 1 0 0 0 0
33 37 1 0 0 0 0
28 31 1 0 0 0 0
16 38 1 0 0 0 0
39 40 1 0 0 0 0
25 39 1 0 0 0 0
41 42 1 0 0 0 0
33 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 0.0000 0.8561
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 0.0000 0.4542
S SKP 5
ID FL5FACGA0002
FORMULA C26H28O16
EXACTMASS 596.137734848
AVERAGEMASS 596.49092
SMILES c(c5O)c(c(c(c5)3)C(C(=C(c(c4)ccc(O)c4O)O3)OC(O2)C(C(O)C(C(CO)2)O)OC(C(O)1)OCC1(O)CO)=O)O
M END