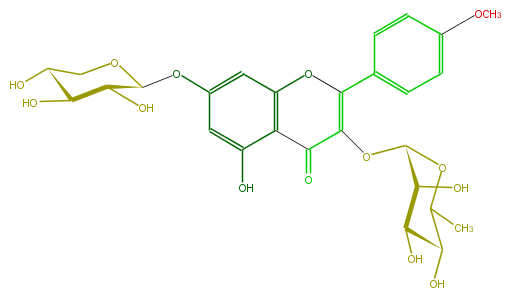
Mol:FL5FABGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3281 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3281 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3281 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3281 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6136 -0.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6136 -0.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1008 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1008 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1008 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1008 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6136 1.6236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6136 1.6236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8152 -0.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8152 -0.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5296 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5296 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5296 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5296 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8152 1.6236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8152 1.6236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8152 -0.6695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8152 -0.6695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2438 1.6235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2438 1.6235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9720 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9720 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7001 1.6235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7001 1.6235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7001 2.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7001 2.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9720 2.8846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9720 2.8846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2438 2.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2438 2.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0422 1.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0422 1.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0862 -0.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0862 -0.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6136 -0.8510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6136 -0.8510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8497 1.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8497 1.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3289 1.0389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3289 1.0389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5792 1.3304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5792 1.3304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7977 1.3218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7977 1.3218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3814 1.8640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3814 1.8640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1473 1.5890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1473 1.5890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4318 1.3900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4318 1.3900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1407 0.9993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1407 0.9993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7770 0.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7770 0.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9218 -1.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9218 -1.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7286 -2.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7286 -2.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4725 -1.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4725 -1.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7286 -0.4108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7286 -0.4108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9218 0.0551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9218 0.0551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1777 -0.8408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1777 -0.8408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0653 -0.8628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0653 -0.8628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0685 -2.3931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0685 -2.3931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5339 -2.9342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5339 -2.9342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0643 -1.7320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0643 -1.7320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5142 2.9342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5142 2.9342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4318 2.4045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4318 2.4045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 15 40 1 0 0 0 0 | + | 15 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 45 -0.8141 -0.4700 | + | M SBV 1 45 -0.8141 -0.4700 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FABGS0005 | + | ID FL5FABGS0005 |
| − | FORMULA C27H30O14 | + | FORMULA C27H30O14 |
| − | EXACTMASS 578.163555668 | + | EXACTMASS 578.163555668 |
| − | AVERAGEMASS 578.5187000000001 | + | AVERAGEMASS 578.5187000000001 |
| − | SMILES c(c1)c(ccc(C(=C(OC(C(O)5)OC(C)C(C5O)O)4)Oc(c2)c(C4=O)c(O)cc(OC(O3)C(O)C(O)C(O)C3)2)1)OC | + | SMILES c(c1)c(ccc(C(=C(OC(C(O)5)OC(C)C(C5O)O)4)Oc(c2)c(C4=O)c(O)cc(OC(O3)C(O)C(O)C(O)C3)2)1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.3281 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3281 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6136 -0.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1008 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1008 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6136 1.6236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8152 -0.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5296 0.3861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5296 1.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8152 1.6236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8152 -0.6695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2438 1.6235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9720 1.2031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7001 1.6235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7001 2.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9720 2.8846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2438 2.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0422 1.6235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0862 -0.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6136 -0.8510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8497 1.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3289 1.0389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5792 1.3304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7977 1.3218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3814 1.8640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1473 1.5890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4318 1.3900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1407 0.9993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7770 0.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9218 -1.7419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7286 -2.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4725 -1.3121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7286 -0.4108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9218 0.0551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1777 -0.8408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0653 -0.8628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0685 -2.3931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5339 -2.9342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0643 -1.7320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5142 2.9342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4318 2.4045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
35 36 1 0 0 0 0
30 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
34 19 1 0 0 0 0
40 41 1 0 0 0 0
15 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 OCH3
M SBV 1 45 -0.8141 -0.4700
S SKP 5
ID FL5FABGS0005
FORMULA C27H30O14
EXACTMASS 578.163555668
AVERAGEMASS 578.5187000000001
SMILES c(c1)c(ccc(C(=C(OC(C(O)5)OC(C)C(C5O)O)4)Oc(c2)c(C4=O)c(O)cc(OC(O3)C(O)C(O)C(O)C3)2)1)OC
M END