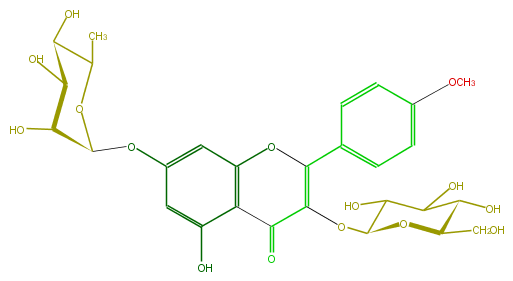
Mol:FL5FABGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.1281 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1281 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1281 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1281 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4136 -1.7709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4136 -1.7709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6991 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6991 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6991 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6991 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4136 -0.1209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4136 -0.1209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0153 -1.7709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0153 -1.7709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7298 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7298 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7298 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7298 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0153 -0.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0153 -0.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0153 -2.4140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0153 -2.4140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4440 -0.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4440 -0.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1722 -0.5414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1722 -0.5414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9003 -0.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9003 -0.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9003 0.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9003 0.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1722 1.1401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1722 1.1401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4440 0.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4440 0.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8423 -0.1210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8423 -0.1210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4528 -1.7720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4528 -1.7720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4136 -2.5955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4136 -2.5955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4523 0.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4523 0.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6455 -0.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6455 -0.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9015 0.6532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9015 0.6532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6455 1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6455 1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4523 2.0204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4523 2.0204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1964 1.1245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1964 1.1245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1218 2.5955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1218 2.5955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6455 2.1357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6455 2.1357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8589 1.6682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8589 1.6682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1119 0.2249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1119 0.2249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3674 -1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3674 -1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9811 -1.9084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9811 -1.9084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7241 -1.6960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7241 -1.6960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4715 -1.9084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4715 -1.9084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8580 -1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8580 -1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1149 -1.4515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1149 -1.4515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7337 -1.3297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7337 -1.3297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7316 -0.9231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7316 -0.9231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4786 -1.3974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4786 -1.3974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1943 -1.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1943 -1.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1119 -2.3639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1119 -2.3639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7145 1.1899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7145 1.1899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6320 0.6601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6320 0.6601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 21 30 1 0 0 0 0 | + | 21 30 1 0 0 0 0 |
| − | 22 18 1 0 0 0 0 | + | 22 18 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 32 19 1 0 0 0 0 | + | 32 19 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 15 42 1 0 0 0 0 | + | 15 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.7227 -0.0743 | + | M SBV 1 45 -0.7227 -0.0743 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 -0.8143 -0.4701 | + | M SBV 2 47 -0.8143 -0.4701 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FABGL0003 | + | ID FL5FABGL0003 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES OC(C1Oc(c5)cc(c(c35)C(C(OC(O4)C(C(O)C(C4CO)O)O)=C(O3)c(c2)ccc(c2)OC)=O)O)C(C(C(O1)C)O)O | + | SMILES OC(C1Oc(c5)cc(c(c35)C(C(OC(O4)C(C(O)C(C4CO)O)O)=C(O3)c(c2)ccc(c2)OC)=O)O)C(C(C(O1)C)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.1281 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1281 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4136 -1.7709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6991 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6991 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4136 -0.1209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0153 -1.7709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7298 -1.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7298 -0.5334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0153 -0.1209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0153 -2.4140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4440 -0.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1722 -0.5414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9003 -0.1210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9003 0.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1722 1.1401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4440 0.7198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8423 -0.1210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4528 -1.7720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4136 -2.5955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4523 0.2233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6455 -0.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9015 0.6532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6455 1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4523 2.0204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1964 1.1245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1218 2.5955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6455 2.1357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8589 1.6682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1119 0.2249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3674 -1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9811 -1.9084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7241 -1.6960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4715 -1.9084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8580 -1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1149 -1.4515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7337 -1.3297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7316 -0.9231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4786 -1.3974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1943 -1.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1119 -2.3639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7145 1.1899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6320 0.6601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
26 29 1 0 0 0 0
21 30 1 0 0 0 0
22 18 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
32 19 1 0 0 0 0
40 41 1 0 0 0 0
34 40 1 0 0 0 0
42 43 1 0 0 0 0
15 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.7227 -0.0743
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 -0.8143 -0.4701
S SKP 5
ID FL5FABGL0003
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES OC(C1Oc(c5)cc(c(c35)C(C(OC(O4)C(C(O)C(C4CO)O)O)=C(O3)c(c2)ccc(c2)OC)=O)O)C(C(C(O1)C)O)O
M END