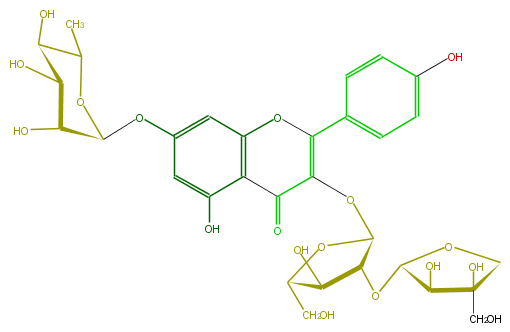
Mol:FL5FAAGS0039
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 49 54 0 0 0 0 0 0 0 0999 V2000 | + | 49 54 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0005 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0005 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0005 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0005 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2860 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2860 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5716 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5716 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5716 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5716 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2860 1.5752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2860 1.5752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1428 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1428 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8572 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8572 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8572 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8572 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1428 1.5752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1428 1.5752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1428 -0.7752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1428 -0.7752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5714 1.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5714 1.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2996 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2996 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0276 1.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0276 1.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0276 2.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0276 2.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2996 2.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2996 2.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5714 2.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5714 2.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2860 -0.7343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2860 -0.7343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7146 1.5750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7146 1.5750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7556 2.8361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7556 2.8361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6609 -0.1350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6609 -0.1350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3708 1.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3708 1.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4709 1.0966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4709 1.0966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9500 1.8956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9500 1.8956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9360 2.8324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9360 2.8324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8359 3.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8359 3.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3568 2.2745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3568 2.2745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7259 3.7330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7259 3.7330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1781 3.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1781 3.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1933 2.6773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1933 2.6773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1005 1.3009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1005 1.3009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3463 -1.8554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3463 -1.8554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0627 -1.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0627 -1.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8671 -1.5375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8671 -1.5375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1330 -0.9426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1330 -0.9426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0609 -1.1070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0609 -1.1070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5600 -1.1805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5600 -1.1805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1765 -2.1357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1765 -2.1357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6907 -1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6907 -1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3109 -2.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3109 -2.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1977 -2.0119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1977 -2.0119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7752 -1.4550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7752 -1.4550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7027 -1.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7027 -1.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3109 -1.5165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3109 -1.5165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1977 -1.5375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1977 -1.5375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7288 -2.5179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7288 -2.5179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1447 -3.7330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1447 -3.7330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1977 -2.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1977 -2.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1933 -2.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1933 -2.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 32 1 0 0 0 0 | + | 36 32 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 39 1 0 0 0 0 | + | 43 39 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | 39 38 1 0 0 0 0 | + | 39 38 1 0 0 0 0 |
| − | 35 21 1 0 0 0 0 | + | 35 21 1 0 0 0 0 |
| − | 41 45 1 0 0 0 0 | + | 41 45 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 32 46 1 0 0 0 0 | + | 32 46 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 41 48 1 0 0 0 0 | + | 41 48 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 46 47 | + | M SAL 1 2 46 47 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 52 -0.3825 0.6625 | + | M SBV 1 52 -0.3825 0.6625 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 48 49 | + | M SAL 2 2 48 49 |
| − | M SBL 2 1 54 | + | M SBL 2 1 54 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 54 0.0000 0.5985 | + | M SBV 2 54 0.0000 0.5985 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGS0039 | + | ID FL5FAAGS0039 |
| − | FORMULA C31H36O18 | + | FORMULA C31H36O18 |
| − | EXACTMASS 696.190164348 | + | EXACTMASS 696.190164348 |
| − | AVERAGEMASS 696.60674 | + | AVERAGEMASS 696.60674 |
| − | SMILES C(OC(=C5c(c6)ccc(c6)O)C(c(c(O5)4)c(O)cc(c4)OC(C3O)OC(C)C(C(O)3)O)=O)(O1)C(OC(O2)C(O)C(O)(C2)CO)C(O)C1CO | + | SMILES C(OC(=C5c(c6)ccc(c6)O)C(c(c(O5)4)c(O)cc(c4)OC(C3O)OC(C)C(C(O)3)O)=O)(O1)C(OC(O2)C(O)C(O)(C2)CO)C(O)C1CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
49 54 0 0 0 0 0 0 0 0999 V2000
-2.0005 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0005 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2860 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5716 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5716 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2860 1.5752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1428 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8572 0.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8572 1.1627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1428 1.5752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1428 -0.7752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5714 1.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2996 1.1546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0276 1.5750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0276 2.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2996 2.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5714 2.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2860 -0.7343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7146 1.5750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7556 2.8361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6609 -0.1350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3708 1.3377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4709 1.0966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9500 1.8956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9360 2.8324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8359 3.0736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3568 2.2745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7259 3.7330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1781 3.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1933 2.6773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1005 1.3009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3463 -1.8554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0627 -1.9707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8671 -1.5375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1330 -0.9426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0609 -1.1070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5600 -1.1805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1765 -2.1357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6907 -1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3109 -2.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1977 -2.0119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7752 -1.4550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7027 -1.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3109 -1.5165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1977 -1.5375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7288 -2.5179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1447 -3.7330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1977 -2.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1933 -2.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
27 30 1 0 0 0 0
22 31 1 0 0 0 0
23 19 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 32 1 0 0 0 0
33 37 1 0 0 0 0
34 38 1 0 0 0 0
39 40 1 1 0 0 0
40 41 1 1 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 39 1 0 0 0 0
40 44 1 0 0 0 0
39 38 1 0 0 0 0
35 21 1 0 0 0 0
41 45 1 0 0 0 0
46 47 1 0 0 0 0
32 46 1 0 0 0 0
48 49 1 0 0 0 0
41 48 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 46 47
M SBL 1 1 52
M SMT 1 CH2OH
M SBV 1 52 -0.3825 0.6625
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 48 49
M SBL 2 1 54
M SMT 2 CH2OH
M SBV 2 54 0.0000 0.5985
S SKP 5
ID FL5FAAGS0039
FORMULA C31H36O18
EXACTMASS 696.190164348
AVERAGEMASS 696.60674
SMILES C(OC(=C5c(c6)ccc(c6)O)C(c(c(O5)4)c(O)cc(c4)OC(C3O)OC(C)C(C(O)3)O)=O)(O1)C(OC(O2)C(O)C(O)(C2)CO)C(O)C1CO
M END