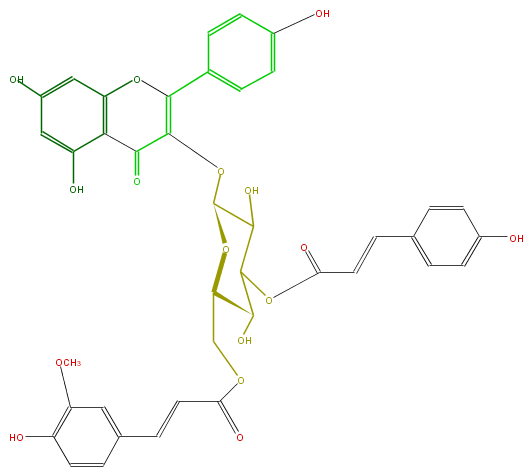
Mol:FL5FAAGL0075
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.9811 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9811 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9811 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9811 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2666 1.9562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2666 1.9562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5522 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5522 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5522 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5522 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2666 3.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2666 3.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8377 1.9562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8377 1.9562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1232 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1232 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1232 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1232 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8377 3.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8377 3.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8377 1.3129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8377 1.3129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2237 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2237 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4955 3.3446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4955 3.3446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2327 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2327 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2327 4.6058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2327 4.6058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4955 5.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4955 5.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2237 4.6058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2237 4.6058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2666 1.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2666 1.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2520 5.0929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2520 5.0929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6168 3.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6168 3.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9336 1.5441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9336 1.5441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9832 -3.6502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9832 -3.6502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5214 -4.4504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5214 -4.4504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9017 -3.6502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9017 -3.6502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3609 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3609 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2145 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2145 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5882 -3.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5882 -3.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3357 -3.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3357 -3.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7094 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7094 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3357 -5.0929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3357 -5.0929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5882 -5.0929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5882 -5.0929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4565 -4.4456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4565 -4.4456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2159 0.2666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2159 0.2666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1093 0.7824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1093 0.7824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8258 -0.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8258 -0.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1093 -1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1093 -1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2159 -1.7231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2159 -1.7231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4994 -0.7313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4994 -0.7313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3269 1.0995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3269 1.0995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1272 -1.3684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1272 -1.3684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4913 -2.2639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4913 -2.2639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1045 -2.3084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1045 -2.3084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4983 -3.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4983 -3.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0621 -0.7542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0621 -0.7542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5213 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5213 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3748 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3748 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7485 -0.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7485 -0.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4961 -0.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4961 -0.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8699 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8699 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4961 0.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4961 0.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7485 0.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7485 0.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6168 0.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6168 0.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2606 -0.7542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2606 -0.7542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9296 -0.1807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9296 -0.1807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5797 -2.7511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5797 -2.7511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6621 -3.2809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6621 -3.2809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 22 24 1 0 0 0 0 | + | 22 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 2 0 0 0 0 | + | 28 29 2 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 29 32 1 0 0 0 0 | + | 29 32 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 34 21 1 0 0 0 0 | + | 34 21 1 0 0 0 0 |
| − | 22 43 1 0 0 0 0 | + | 22 43 1 0 0 0 0 |
| − | 43 42 1 0 0 0 0 | + | 43 42 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 49 52 1 0 0 0 0 | + | 49 52 1 0 0 0 0 |
| − | 44 53 1 0 0 0 0 | + | 44 53 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 40 53 1 0 0 0 0 | + | 40 53 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 28 55 1 0 0 0 0 | + | 28 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 55 56 | + | M SAL 1 2 55 56 |
| − | M SBL 1 1 61 | + | M SBL 1 1 61 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 61 0.2440 -1.0472 | + | M SBV 1 61 0.2440 -1.0472 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0075 | + | ID FL5FAAGL0075 |
| − | FORMULA C40H34O16 | + | FORMULA C40H34O16 |
| − | EXACTMASS 770.18468504 | + | EXACTMASS 770.18468504 |
| − | AVERAGEMASS 770.68836 | + | AVERAGEMASS 770.68836 |
| − | SMILES c(c1O)cc(C=CC(OC(C5O)C(C(OC5COC(C=Cc(c6)cc(OC)c(O)c6)=O)OC(C(=O)2)=C(c(c4)ccc(O)c4)Oc(c3)c2c(cc(O)3)O)O)=O)cc1 | + | SMILES c(c1O)cc(C=CC(OC(C5O)C(C(OC5COC(C=Cc(c6)cc(OC)c(O)c6)=O)OC(C(=O)2)=C(c(c4)ccc(O)c4)Oc(c3)c2c(cc(O)3)O)O)=O)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-4.9811 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9811 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2666 1.9562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5522 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5522 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2666 3.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8377 1.9562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1232 2.3688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1232 3.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8377 3.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8377 1.3129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2237 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4955 3.3446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2327 3.7649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2327 4.6058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4955 5.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2237 4.6058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2666 1.1316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2520 5.0929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.6168 3.6062 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9336 1.5441 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9832 -3.6502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5214 -4.4504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9017 -3.6502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3609 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2145 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5882 -3.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3357 -3.7983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7094 -4.4456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3357 -5.0929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5882 -5.0929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4565 -4.4456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2159 0.2666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1093 0.7824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8258 -0.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1093 -1.2074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2159 -1.7231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4994 -0.7313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3269 1.0995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1272 -1.3684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4913 -2.2639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1045 -2.3084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4983 -3.1653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0621 -0.7542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5213 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3748 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7485 -0.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4961 -0.6061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8699 0.0412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4961 0.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7485 0.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6168 0.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2606 -0.7542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9296 -0.1807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5797 -2.7511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6621 -3.2809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
22 23 2 0 0 0 0
22 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 28 1 0 0 0 0
28 29 2 0 0 0 0
29 30 1 0 0 0 0
30 31 2 0 0 0 0
31 26 1 0 0 0 0
29 32 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
38 40 1 0 0 0 0
37 41 1 0 0 0 0
36 42 1 0 0 0 0
34 21 1 0 0 0 0
22 43 1 0 0 0 0
43 42 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
49 52 1 0 0 0 0
44 53 1 0 0 0 0
53 54 2 0 0 0 0
40 53 1 0 0 0 0
4 3 1 0 0 0 0
55 56 1 0 0 0 0
28 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 55 56
M SBL 1 1 61
M SMT 1 ^OCH3
M SBV 1 61 0.2440 -1.0472
S SKP 5
ID FL5FAAGL0075
FORMULA C40H34O16
EXACTMASS 770.18468504
AVERAGEMASS 770.68836
SMILES c(c1O)cc(C=CC(OC(C5O)C(C(OC5COC(C=Cc(c6)cc(OC)c(O)c6)=O)OC(C(=O)2)=C(c(c4)ccc(O)c4)Oc(c3)c2c(cc(O)3)O)O)=O)cc1
M END