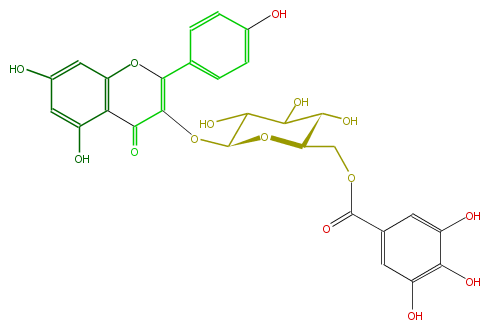
Mol:FL5FAAGL0065
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.8768 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8768 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8768 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8768 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3205 0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3205 0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7642 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7642 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7642 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7642 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3205 2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3205 2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2079 0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2079 0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6516 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6516 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6516 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6516 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2079 2.0359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2079 2.0359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2079 0.2504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2079 0.2504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0955 2.0358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0955 2.0358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5286 1.7085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5286 1.7085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0384 2.0358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0384 2.0358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0384 2.6905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0384 2.6905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5286 3.0179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5286 3.0179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0955 2.6905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0955 2.6905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0035 0.5221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0035 0.5221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0419 1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0419 1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3457 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3457 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3997 0.5551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3997 0.5551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1497 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1497 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5374 1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5374 1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7919 0.8005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7919 0.8005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6852 0.8187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6852 0.8187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0607 1.2661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0607 1.2661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0237 0.8832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0237 0.8832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6052 3.0178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6052 3.0178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3205 0.1091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3205 0.1091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4878 1.9133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4878 1.9133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7786 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7786 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1242 -0.2565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1242 -0.2565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1242 -0.9871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1242 -0.9871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6239 -1.2760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6239 -1.2760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7565 -1.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7565 -1.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7565 -2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7565 -2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3340 -2.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3340 -2.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9114 -2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9114 -2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9114 -1.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9114 -1.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3340 -1.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3340 -1.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4878 -1.0194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4878 -1.0194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4878 -2.3517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4878 -2.3517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3340 -3.0179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3340 -3.0179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGL0065 | + | ID FL5FAAGL0065 |
| − | KNApSAcK_ID C00005847 | + | KNApSAcK_ID C00005847 |
| − | NAME Kaempferol 3-(6''-galloylglucoside) | + | NAME Kaempferol 3-(6''-galloylglucoside) |
| − | CAS_RN 56317-05-6 | + | CAS_RN 56317-05-6 |
| − | FORMULA C28H24O15 | + | FORMULA C28H24O15 |
| − | EXACTMASS 600.111520098 | + | EXACTMASS 600.111520098 |
| − | AVERAGEMASS 600.48116 | + | AVERAGEMASS 600.48116 |
| − | SMILES Oc(c1)c(c(O)cc1C(OCC(C(O)5)OC(C(O)C5O)OC(=C3c(c4)ccc(O)c4)C(c(c(O3)2)c(cc(O)c2)O)=O)=O)O | + | SMILES Oc(c1)c(c(O)cc1C(OCC(C(O)5)OC(C(O)C5O)OC(=C3c(c4)ccc(O)c4)C(c(c(O3)2)c(cc(O)c2)O)=O)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.8768 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8768 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3205 0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7642 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7642 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3205 2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2079 0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6516 1.0724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6516 1.7148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2079 2.0359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2079 0.2504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0955 2.0358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5286 1.7085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0384 2.0358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0384 2.6905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5286 3.0179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0955 2.6905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0035 0.5221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0419 1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3457 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3997 0.5551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1497 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5374 1.0135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7919 0.8005 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6852 0.8187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0607 1.2661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0237 0.8832 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6052 3.0178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3205 0.1091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4878 1.9133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7786 0.3421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1242 -0.2565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1242 -0.9871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6239 -1.2760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7565 -1.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7565 -2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3340 -2.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9114 -2.0190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9114 -1.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3340 -1.0188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4878 -1.0194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4878 -2.3517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3340 -3.0179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
39 41 1 0 0 0 0
38 42 1 0 0 0 0
37 43 1 0 0 0 0
S SKP 8
ID FL5FAAGL0065
KNApSAcK_ID C00005847
NAME Kaempferol 3-(6''-galloylglucoside)
CAS_RN 56317-05-6
FORMULA C28H24O15
EXACTMASS 600.111520098
AVERAGEMASS 600.48116
SMILES Oc(c1)c(c(O)cc1C(OCC(C(O)5)OC(C(O)C5O)OC(=C3c(c4)ccc(O)c4)C(c(c(O3)2)c(cc(O)c2)O)=O)=O)O
M END