Mol:FL5FAAGL0019
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.3329 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3329 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3329 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3329 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6319 -0.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6319 -0.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9309 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9309 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9309 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9309 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6319 1.1282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6319 1.1282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2298 -0.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2298 -0.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5287 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5287 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5287 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5287 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2298 1.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2298 1.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2298 -1.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2298 -1.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8281 1.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8281 1.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1136 0.7155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1136 0.7155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6009 1.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6009 1.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6009 1.9531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6009 1.9531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1136 2.3655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1136 2.3655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8281 1.9531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8281 1.9531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6319 -1.3000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6319 -1.3000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0337 1.1280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0337 1.1280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3152 2.3654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3152 2.3654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6861 -0.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6861 -0.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8001 -0.5656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8001 -0.5656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0517 -1.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0517 -1.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0264 -1.2627 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0264 -1.2627 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8225 -1.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8225 -1.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5711 -1.2102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5711 -1.2102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5964 -1.1411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5964 -1.1411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0433 -0.4187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0433 -0.4187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3238 -0.7602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3238 -0.7602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1649 -1.5769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1649 -1.5769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7102 2.6720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7102 2.6720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9618 2.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9618 2.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9364 1.9750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9364 1.9750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7327 1.3995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7327 1.3995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4811 2.0275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4811 2.0275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5065 2.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5065 2.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9515 2.7538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9515 2.7538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2113 2.4729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2113 2.4729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9805 1.6589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9805 1.6589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3654 1.1977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3654 1.1977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0337 0.4013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0337 0.4013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4855 -1.9574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4855 -1.9574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1538 -2.7538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1538 -2.7538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.6328 0.2019 | + | M SBV 1 45 -0.6328 0.2019 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 47 -0.6630 0.1192 | + | M SBV 2 47 -0.6630 0.1192 |
| − | S SKP 5 | + | S SKP 5 |
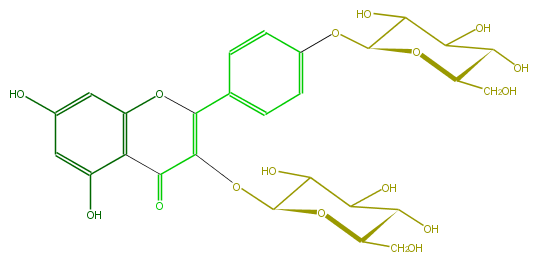
| − | ID FL5FAAGL0019 | + | ID FL5FAAGL0019 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(c1)(O3)c(C(C(=C(c(c4)ccc(OC(O5)C(O)C(O)C(O)C5CO)c4)3)OC(C(O)2)OC(C(C(O)2)O)CO)=O)c(O)cc(O)1 | + | SMILES c(c1)(O3)c(C(C(=C(c(c4)ccc(OC(O5)C(O)C(O)C(O)C5CO)c4)3)OC(C(O)2)OC(C(C(O)2)O)CO)=O)c(O)cc(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-4.3329 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3329 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6319 -0.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9309 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9309 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6319 1.1282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2298 -0.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5287 -0.0861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5287 0.7234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2298 1.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2298 -1.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8281 1.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1136 0.7155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6009 1.1280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6009 1.9531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1136 2.3655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8281 1.9531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6319 -1.3000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0337 1.1280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3152 2.3654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6861 -0.7275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8001 -0.5656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0517 -1.1937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0264 -1.2627 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8225 -1.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5711 -1.2102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5964 -1.1411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0433 -0.4187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3238 -0.7602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1649 -1.5769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7102 2.6720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9618 2.0441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9364 1.9750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7327 1.3995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4811 2.0275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5065 2.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9515 2.7538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2113 2.4729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9805 1.6589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3654 1.1977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0337 0.4013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4855 -1.9574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1538 -2.7538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
32 20 1 0 0 0 0
40 41 1 0 0 0 0
34 40 1 0 0 0 0
42 43 1 0 0 0 0
25 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.6328 0.2019
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 CH2OH
M SBV 2 47 -0.6630 0.1192
S SKP 5
ID FL5FAAGL0019
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(c1)(O3)c(C(C(=C(c(c4)ccc(OC(O5)C(O)C(O)C(O)C5CO)c4)3)OC(C(O)2)OC(C(C(O)2)O)CO)=O)c(O)cc(O)1
M END