Mol:FL5F4GGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.4746 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4746 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7602 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7602 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7602 1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7602 1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4746 1.4435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4746 1.4435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1891 1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1891 1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1891 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1891 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0457 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0457 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3313 0.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3313 0.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6168 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6168 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6168 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6168 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3313 -1.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3313 -1.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0457 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0457 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0976 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0976 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8120 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8120 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8120 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8120 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0976 -1.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0976 -1.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3313 -2.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3313 -2.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5265 0.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5265 0.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8213 -1.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8213 -1.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9035 1.4435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9035 1.4435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0976 0.9026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0976 0.9026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9061 -0.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9061 -0.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4748 -1.4140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4748 -1.4140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4746 2.1230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4746 2.1230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1491 2.5124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1491 2.5124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3212 0.6347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3212 0.6347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9086 -0.0800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9086 -0.0800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1102 0.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1102 0.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3167 -0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3167 -0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7293 0.6170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7293 0.6170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5277 0.4079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5277 0.4079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8483 0.9099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8483 0.9099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4725 0.9188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4725 0.9188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9061 0.3976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9061 0.3976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5414 0.0126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5414 0.0126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5791 -0.5121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5791 -0.5121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0325 -1.2204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0325 -1.2204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4451 -1.9351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4451 -1.9351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6516 -1.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6516 -1.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8534 -1.9176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8534 -1.9176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4407 -1.2029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4407 -1.2029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2341 -1.4296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2341 -1.4296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7961 -2.5124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7961 -2.5124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1675 -2.3082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1675 -2.3082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8964 -1.6745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8964 -1.6745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6446 -1.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6446 -1.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 6 22 1 0 0 0 0 | + | 6 22 1 0 0 0 0 |
| − | 15 23 1 0 0 0 0 | + | 15 23 1 0 0 0 0 |
| − | 4 24 1 0 0 0 0 | + | 4 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 26 34 1 0 0 0 0 | + | 26 34 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 28 36 1 0 0 0 0 | + | 28 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 39 44 1 0 0 0 0 | + | 39 44 1 0 0 0 0 |
| − | 38 45 1 0 0 0 0 | + | 38 45 1 0 0 0 0 |
| − | 37 46 1 0 0 0 0 | + | 37 46 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 29 18 1 0 0 0 0 | + | 29 18 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5F4GGS0001 | + | ID FL5F4GGS0001 |
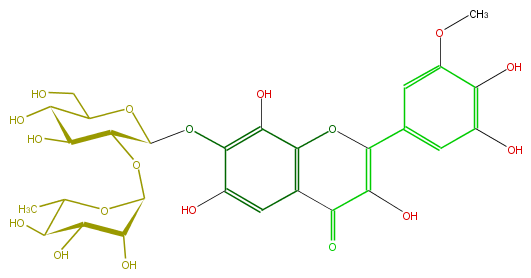
| − | FORMULA C28H32O18 | + | FORMULA C28H32O18 |
| − | EXACTMASS 656.1588642199999 | + | EXACTMASS 656.1588642199999 |
| − | AVERAGEMASS 656.54288 | + | AVERAGEMASS 656.54288 |
| − | SMILES C(O)(C(CO)2)C(O)C(C(Oc(c(O)3)c(cc(C(=O)5)c3OC(=C5O)c(c4)cc(OC)c(c4O)O)O)O2)OC(C1O)OC(C)C(C1O)O | + | SMILES C(O)(C(CO)2)C(O)C(C(Oc(c(O)3)c(cc(C(=O)5)c3OC(=C5O)c(c4)cc(OC)c(c4O)O)O)O2)OC(C1O)OC(C)C(C1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
3.4746 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7602 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7602 1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4746 1.4435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1891 1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1891 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0457 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3313 0.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6168 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6168 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3313 -1.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0457 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0976 0.2060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8120 -0.2065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8120 -1.0314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0976 -1.4439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3313 -2.1508 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5265 0.2060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8213 -1.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9035 1.4435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0976 0.9026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9061 -0.2080 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4748 -1.4140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4746 2.1230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1491 2.5124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3212 0.6347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9086 -0.0800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1102 0.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3167 -0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7293 0.6170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5277 0.4079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8483 0.9099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4725 0.9188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9061 0.3976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5414 0.0126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5791 -0.5121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0325 -1.2204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4451 -1.9351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6516 -1.7083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8534 -1.9176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4407 -1.2029 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2341 -1.4296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7961 -2.5124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1675 -2.3082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8964 -1.6745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6446 -1.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
5 20 1 0 0 0 0
13 21 1 0 0 0 0
6 22 1 0 0 0 0
15 23 1 0 0 0 0
4 24 1 0 0 0 0
24 25 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
26 34 1 0 0 0 0
27 35 1 0 0 0 0
28 36 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 1 0 0 0
40 39 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
39 44 1 0 0 0 0
38 45 1 0 0 0 0
37 46 1 0 0 0 0
41 36 1 0 0 0 0
29 18 1 0 0 0 0
S SKP 5
ID FL5F4GGS0001
FORMULA C28H32O18
EXACTMASS 656.1588642199999
AVERAGEMASS 656.54288
SMILES C(O)(C(CO)2)C(O)C(C(Oc(c(O)3)c(cc(C(=O)5)c3OC(=C5O)c(c4)cc(OC)c(c4O)O)O)O2)OC(C1O)OC(C)C(C1O)O
M END