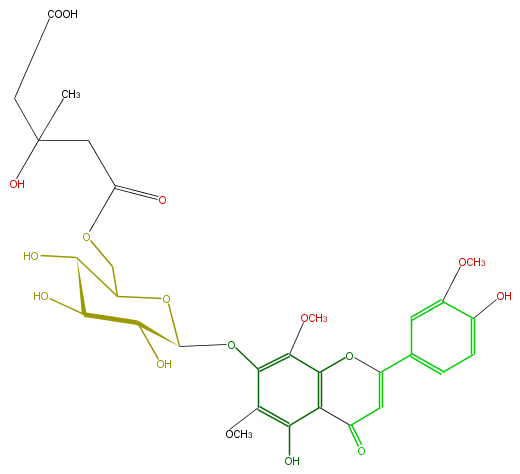
Mol:FL3FGCGS0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 50 0 0 0 0 0 0 0 0999 V2000 | + | 47 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0303 -3.1181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0303 -3.1181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0303 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0303 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6924 -4.3252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6924 -4.3252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3545 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3545 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3545 -3.1785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3545 -3.1785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6924 -2.7963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6924 -2.7963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0164 -4.3252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0164 -4.3252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6785 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6785 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6785 -3.1785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6785 -3.1785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0164 -2.7963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0164 -2.7963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2684 -4.8652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2684 -4.8652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3403 -2.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3403 -2.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0150 -3.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0150 -3.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6898 -2.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6898 -2.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6898 -2.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6898 -2.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0150 -1.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0150 -1.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3403 -2.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3403 -2.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6924 -5.0882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6924 -5.0882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5480 -2.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5480 -2.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2799 -1.5140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2799 -1.5140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9141 -0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9141 -0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7580 -1.9214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7580 -1.9214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6024 -2.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6024 -2.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6860 -2.6152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6860 -2.6152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9658 -1.5713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9658 -1.5713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0429 -1.5303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0429 -1.5303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1444 -0.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1444 -0.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8140 -0.6352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8140 -0.6352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6089 -1.5148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6089 -1.5148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0998 -2.9906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0998 -2.9906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7004 -0.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7004 -0.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0798 0.8492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0798 0.8492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0479 0.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0479 0.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6581 1.8511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6581 1.8511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7467 1.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7467 1.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1576 2.8717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1576 2.8717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2787 0.9298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2787 0.9298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2799 2.7748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2799 2.7748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4536 -0.7691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4536 -0.7691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1213 0.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1213 0.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4690 4.6050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4690 4.6050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8112 4.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8112 4.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9426 5.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9426 5.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0428 -1.9884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0428 -1.9884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6270 -0.6419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6270 -0.6419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7026 -4.4911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7026 -4.4911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8347 -5.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8347 -5.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 31 27 1 0 0 0 0 | + | 31 27 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 16 39 1 0 0 0 0 | + | 16 39 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 6 44 1 0 0 0 0 | + | 6 44 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 2 46 1 0 0 0 0 | + | 2 46 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 43 -0.4386 -0.8587 | + | M SBV 1 43 -0.4386 -0.8587 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 41 42 43 | + | M SAL 2 3 41 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 46 -0.8109 -1.8302 | + | M SBV 2 46 -0.8109 -1.8302 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 48 -0.3504 -0.8078 | + | M SBV 3 48 -0.3504 -0.8078 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 46 47 | + | M SAL 4 2 46 47 |
| − | M SBL 4 1 50 | + | M SBL 4 1 50 |
| − | M SMT 4 ^ OCH3 | + | M SMT 4 ^ OCH3 |
| − | M SBV 4 50 0.7329 0.5480 | + | M SBV 4 50 0.7329 0.5480 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FGCGS0008 | + | ID FL3FGCGS0008 |
| − | FORMULA C30H34O17 | + | FORMULA C30H34O17 |
| − | EXACTMASS 666.179599662 | + | EXACTMASS 666.179599662 |
| − | AVERAGEMASS 666.58076 | + | AVERAGEMASS 666.58076 |
| − | SMILES OC(C1O)C(C(OC(Oc(c(OC)2)c(c(O)c(C4=O)c(OC(=C4)c(c3)ccc(O)c3OC)2)OC)1)COC(=O)CC(CC(O)=O)(C)O)O | + | SMILES OC(C1O)C(C(OC(Oc(c(OC)2)c(c(O)c(C4=O)c(OC(=C4)c(c3)ccc(O)c3OC)2)OC)1)COC(=O)CC(CC(O)=O)(C)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 50 0 0 0 0 0 0 0 0999 V2000
0.0303 -3.1181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0303 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6924 -4.3252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3545 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3545 -3.1785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6924 -2.7963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0164 -4.3252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6785 -3.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6785 -3.1785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0164 -2.7963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2684 -4.8652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3403 -2.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0150 -3.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6898 -2.7965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6898 -2.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0150 -1.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3403 -2.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6924 -5.0882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5480 -2.5706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2799 -1.5140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9141 -0.6778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7580 -1.9214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6024 -2.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6860 -2.6152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9658 -1.5713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0429 -1.5303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1444 -0.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8140 -0.6352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6089 -1.5148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0998 -2.9906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7004 -0.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0798 0.8492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0479 0.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6581 1.8511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7467 1.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1576 2.8717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2787 0.9298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2799 2.7748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4536 -0.7691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1213 0.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4690 4.6050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8112 4.0121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9426 5.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0428 -1.9884 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6270 -0.6419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7026 -4.4911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8347 -5.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
31 27 1 0 0 0 0
24 19 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
35 37 1 0 0 0 0
35 38 1 0 0 0 0
39 40 1 0 0 0 0
16 39 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
38 41 1 0 0 0 0
44 45 1 0 0 0 0
6 44 1 0 0 0 0
46 47 1 0 0 0 0
2 46 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 OCH3
M SBV 1 43 -0.4386 -0.8587
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 41 42 43
M SBL 2 1 46
M SMT 2 COOH
M SBV 2 46 -0.8109 -1.8302
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 48
M SMT 3 OCH3
M SBV 3 48 -0.3504 -0.8078
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 46 47
M SBL 4 1 50
M SMT 4 ^ OCH3
M SBV 4 50 0.7329 0.5480
S SKP 5
ID FL3FGCGS0008
FORMULA C30H34O17
EXACTMASS 666.179599662
AVERAGEMASS 666.58076
SMILES OC(C1O)C(C(OC(Oc(c(OC)2)c(c(O)c(C4=O)c(OC(=C4)c(c3)ccc(O)c3OC)2)OC)1)COC(=O)CC(CC(O)=O)(C)O)O
M END