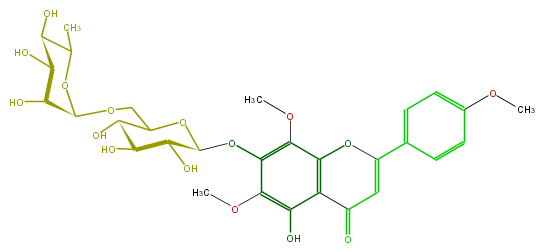
Mol:FL3FGAGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.3527 0.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3527 0.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3527 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3527 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0672 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0672 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7817 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7817 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7817 0.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7817 0.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0672 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0672 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6383 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6383 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9238 -0.4126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9238 -0.4126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2093 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2093 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2093 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2093 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9238 -2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9238 -2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6383 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6383 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4949 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4949 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2196 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2196 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2196 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2196 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4949 -2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4949 -2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9238 -2.7808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9238 -2.7808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9341 -0.4126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9341 -0.4126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4949 -2.7486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4949 -2.7486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4873 0.8139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4873 0.8139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4949 0.2418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4949 0.2418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8696 -2.0254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8696 -2.0254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5663 -1.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5663 -1.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1557 0.4280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1557 0.4280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2670 0.6816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2670 0.6816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7294 0.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7294 0.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3169 -0.5912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3169 -0.5912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5188 -0.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5188 -0.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7256 -0.6090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7256 -0.6090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1380 0.1056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1380 0.1056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9361 -0.1034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9361 -0.1034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0322 -1.0397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0322 -1.0397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2726 -0.2165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2726 -0.2165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8867 -0.5607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8867 -0.5607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4064 0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4064 0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9223 0.3905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9223 0.3905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8863 1.8235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8863 1.8235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6220 2.1969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6220 2.1969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3528 1.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3528 1.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5187 0.6088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5187 0.6088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7830 0.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7830 0.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0521 1.0153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0521 1.0153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4787 2.7808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4787 2.7808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0084 2.4499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0084 2.4499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0226 1.8273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0226 1.8273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1557 0.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1557 0.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 15 22 1 0 0 0 0 | + | 15 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 20 24 1 0 0 0 0 | + | 20 24 1 0 0 0 0 |
| − | 21 25 1 0 0 0 0 | + | 21 25 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 26 33 1 0 0 0 0 | + | 26 33 1 0 0 0 0 |
| − | 27 34 1 0 0 0 0 | + | 27 34 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 38 43 1 0 0 0 0 | + | 38 43 1 0 0 0 0 |
| − | 37 44 1 0 0 0 0 | + | 37 44 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 36 41 1 0 0 0 0 | + | 36 41 1 0 0 0 0 |
| − | 29 18 1 0 0 0 0 | + | 29 18 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FGAGS0005 | + | ID FL3FGAGS0005 |
| − | FORMULA C30H36O16 | + | FORMULA C30H36O16 |
| − | EXACTMASS 652.200335104 | + | EXACTMASS 652.200335104 |
| − | AVERAGEMASS 652.59724 | + | AVERAGEMASS 652.59724 |
| − | SMILES C(Oc(c5OC)c(OC)c(O3)c(c5O)C(=O)C=C(c(c4)ccc(c4)OC)3)(O1)C(O)C(C(O)C(COC(C2O)OC(C(C(O)2)O)C)1)O | + | SMILES C(Oc(c5OC)c(OC)c(O3)c(c5O)C(=O)C=C(c(c4)ccc(c4)OC)3)(O1)C(O)C(C(O)C(COC(C2O)OC(C(C(O)2)O)C)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
3.3527 0.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3527 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0672 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7817 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7817 0.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0672 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6383 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9238 -0.4126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2093 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2093 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9238 -2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6383 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4949 -0.4126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2196 -0.8251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2196 -1.6501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4949 -2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9238 -2.7808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9341 -0.4126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4949 -2.7486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4873 0.8139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4949 0.2418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8696 -2.0254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5663 -1.6232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1557 0.4280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2670 0.6816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7294 0.1232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3169 -0.5912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5188 -0.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7256 -0.6090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1380 0.1056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9361 -0.1034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0322 -1.0397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2726 -0.2165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8867 -0.5607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4064 0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9223 0.3905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8863 1.8235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6220 2.1969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3528 1.4170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5187 0.6088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7830 0.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0521 1.0153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4787 2.7808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0084 2.4499 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0226 1.8273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.1557 0.5930 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
16 19 1 0 0 0 0
5 20 1 0 0 0 0
13 21 1 0 0 0 0
15 22 1 0 0 0 0
22 23 1 0 0 0 0
20 24 1 0 0 0 0
21 25 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
28 32 1 0 0 0 0
26 33 1 0 0 0 0
27 34 1 0 0 0 0
31 35 1 0 0 0 0
35 36 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 1 0 0 0
40 39 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 37 1 0 0 0 0
38 43 1 0 0 0 0
37 44 1 0 0 0 0
39 45 1 0 0 0 0
40 46 1 0 0 0 0
36 41 1 0 0 0 0
29 18 1 0 0 0 0
S SKP 5
ID FL3FGAGS0005
FORMULA C30H36O16
EXACTMASS 652.200335104
AVERAGEMASS 652.59724
SMILES C(Oc(c5OC)c(OC)c(O3)c(c5O)C(=O)C=C(c(c4)ccc(c4)OC)3)(O1)C(O)C(C(O)C(COC(C2O)OC(C(C(O)2)O)C)1)O
M END