Mol:FL3FF8GS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 40 0 0 0 0 0 0 0 0999 V2000 | + | 37 40 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5027 0.6369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5027 0.6369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5027 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5027 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0517 -0.1856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0517 -0.1856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3994 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3994 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3994 0.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3994 0.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0517 0.8561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0517 0.8561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8505 -0.1856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8505 -0.1856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3015 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3015 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3015 0.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3015 0.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8505 0.8561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8505 0.8561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0222 -0.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0222 -0.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7524 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7524 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2122 0.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2122 0.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6719 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6719 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6719 1.3868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6719 1.3868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2122 1.6522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2122 1.6522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7524 1.3868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7524 1.3868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0517 -0.7054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0517 -0.7054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6204 -1.1147 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.6204 -1.1147 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.1048 -1.7953 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1048 -1.7953 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.3623 -1.5066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.3623 -1.5066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6458 -1.4989 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6458 -1.4989 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.1665 -0.9781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1665 -0.9781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9249 -1.2505 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.9249 -1.2505 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.1969 -1.4476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1969 -1.4476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9086 -1.8345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9086 -1.8345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9369 -2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9369 -2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4825 2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4825 2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1969 1.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1969 1.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8437 1.2837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8437 1.2837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3101 2.1683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3101 2.1683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3952 2.0056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3952 2.0056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8953 2.8717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8953 2.8717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1872 1.4064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1872 1.4064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5854 2.3237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5854 2.3237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5340 -0.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5340 -0.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9466 0.0708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9466 0.0708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 18 1 0 0 0 0 | + | 22 18 1 0 0 0 0 |
| − | 16 28 1 0 0 0 0 | + | 16 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 17 32 1 0 0 0 0 | + | 17 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 6 34 1 0 0 0 0 | + | 6 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 24 36 1 0 0 0 0 | + | 24 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 36 37 | + | M SAL 5 2 36 37 |
| − | M SBL 5 1 39 | + | M SBL 5 1 39 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 39 -2.2205 -0.4431 | + | M SVB 5 39 -2.2205 -0.4431 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 34 35 | + | M SAL 4 2 34 35 |
| − | M SBL 4 1 37 | + | M SBL 4 1 37 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 37 0.1872 1.4064 | + | M SVB 4 37 0.1872 1.4064 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 32 33 | + | M SAL 3 2 32 33 |
| − | M SBL 3 1 35 | + | M SBL 3 1 35 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 35 1.3952 2.0056 | + | M SVB 3 35 1.3952 2.0056 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 30 31 | + | M SAL 2 2 30 31 |
| − | M SBL 2 1 33 | + | M SBL 2 1 33 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 33 -0.8437 1.2837 | + | M SVB 2 33 -0.8437 1.2837 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 28 29 | + | M SAL 1 2 28 29 |
| − | M SBL 1 1 31 | + | M SBL 1 1 31 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 31 2.4825 2.2208 | + | M SVB 1 31 2.4825 2.2208 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FF8GS0005 | + | ID FL3FF8GS0005 |
| − | KNApSAcK_ID C00004450 | + | KNApSAcK_ID C00004450 |
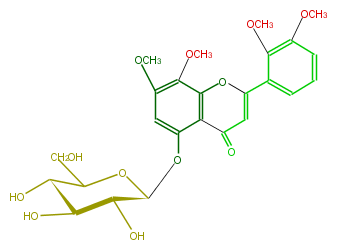
| − | NAME 5-Hydroxy-7,8,2',3'-tetramethoxyflavone 5-glucoside | + | NAME 5-Hydroxy-7,8,2',3'-tetramethoxyflavone 5-glucoside |
| − | CAS_RN 113963-40-9 | + | CAS_RN 113963-40-9 |
| − | FORMULA C25H28O12 | + | FORMULA C25H28O12 |
| − | EXACTMASS 520.15807636 | + | EXACTMASS 520.15807636 |
| − | AVERAGEMASS 520.48262 | + | AVERAGEMASS 520.48262 |
| − | SMILES c(c4OC)(c(O1)c(c(c4)O[C@@H]([C@H]3O)OC(CO)[C@H](O)[C@@H]3O)C(C=C(c(c2)c(c(cc2)OC)OC)1)=O)OC | + | SMILES c(c4OC)(c(O1)c(c(c4)O[C@@H]([C@H]3O)OC(CO)[C@H](O)[C@@H]3O)C(C=C(c(c2)c(c(cc2)OC)OC)1)=O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 40 0 0 0 0 0 0 0 0999 V2000
-0.5027 0.6369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5027 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0517 -0.1856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3994 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3994 0.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0517 0.8561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8505 -0.1856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3015 0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3015 0.5956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8505 0.8561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0222 -0.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7524 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2122 0.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6719 0.8560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6719 1.3868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2122 1.6522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7524 1.3868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0517 -0.7054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6204 -1.1147 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.1048 -1.7953 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3623 -1.5066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6458 -1.4989 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.1665 -0.9781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9249 -1.2505 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.1969 -1.4476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9086 -1.8345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9369 -2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4825 2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1969 1.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8437 1.2837 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3101 2.1683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3952 2.0056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8953 2.8717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1872 1.4064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5854 2.3237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5340 -0.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9466 0.0708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 18 1 0 0 0 0
16 28 1 0 0 0 0
28 29 1 0 0 0 0
1 30 1 0 0 0 0
30 31 1 0 0 0 0
17 32 1 0 0 0 0
32 33 1 0 0 0 0
6 34 1 0 0 0 0
34 35 1 0 0 0 0
24 36 1 0 0 0 0
36 37 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 36 37
M SBL 5 1 39
M SMT 5 CH2OH
M SVB 5 39 -2.2205 -0.4431
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 34 35
M SBL 4 1 37
M SMT 4 OCH3
M SVB 4 37 0.1872 1.4064
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 32 33
M SBL 3 1 35
M SMT 3 OCH3
M SVB 3 35 1.3952 2.0056
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 30 31
M SBL 2 1 33
M SMT 2 OCH3
M SVB 2 33 -0.8437 1.2837
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 28 29
M SBL 1 1 31
M SMT 1 OCH3
M SVB 1 31 2.4825 2.2208
S SKP 8
ID FL3FF8GS0005
KNApSAcK_ID C00004450
NAME 5-Hydroxy-7,8,2',3'-tetramethoxyflavone 5-glucoside
CAS_RN 113963-40-9
FORMULA C25H28O12
EXACTMASS 520.15807636
AVERAGEMASS 520.48262
SMILES c(c4OC)(c(O1)c(c(c4)O[C@@H]([C@H]3O)OC(CO)[C@H](O)[C@@H]3O)C(C=C(c(c2)c(c(cc2)OC)OC)1)=O)OC
M END