Mol:FL3FCADS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9610 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9610 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9610 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9610 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2466 -2.3150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2466 -2.3150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5322 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5322 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5322 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5322 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2466 -0.6651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2466 -0.6651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8177 -2.3150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8177 -2.3150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1033 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1033 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1033 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1033 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8177 -0.6651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8177 -0.6651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8177 -2.9581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8177 -2.9581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1229 2.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1229 2.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9006 1.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9006 1.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5707 0.9149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5707 0.9149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5619 0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5619 0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9669 0.6912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9669 0.6912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3586 1.4335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3586 1.4335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4489 3.1396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4489 3.1396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9027 2.5362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9027 2.5362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9316 0.2831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9316 0.2831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2466 -3.1396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2466 -3.1396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2303 -0.5327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2303 -0.5327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5227 -0.9674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5227 -0.9674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2755 -0.5327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2755 -0.5327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2755 0.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2755 0.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5227 0.7712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5227 0.7712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2303 0.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2303 0.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8422 0.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8422 0.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9301 1.3007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9301 1.3007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5438 0.6317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5438 0.6317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2866 0.8439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2866 0.8439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0341 0.6317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0341 0.6317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4205 1.3007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4205 1.3007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6776 1.0886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6776 1.0886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2759 1.2721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2759 1.2721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1242 1.5935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1242 1.5935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0619 1.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0619 1.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4198 -0.2829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4198 -0.2829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0619 0.8292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0619 0.8292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9174 1.9480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9174 1.9480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9998 1.4183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9998 1.4183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7868 0.6673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7868 0.6673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7868 -0.3921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7868 -0.3921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 12 13 1 1 0 0 0 | + | 12 13 1 1 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 15 14 1 1 0 0 0 | + | 15 14 1 1 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 12 18 1 0 0 0 0 | + | 12 18 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 6 15 1 0 0 0 0 | + | 6 15 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 9 22 1 0 0 0 0 | + | 9 22 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 1 38 1 0 0 0 0 | + | 1 38 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 17 40 1 0 0 0 0 | + | 17 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 32 42 1 0 0 0 0 | + | 32 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 43 0.4587 -0.7946 | + | M SBV 1 43 0.4587 -0.7946 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 45 -0.4412 -0.5145 | + | M SBV 2 45 -0.4412 -0.5145 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 47 -0.7527 -0.0356 | + | M SBV 3 47 -0.7527 -0.0356 |
| − | S SKP 5 | + | S SKP 5 |
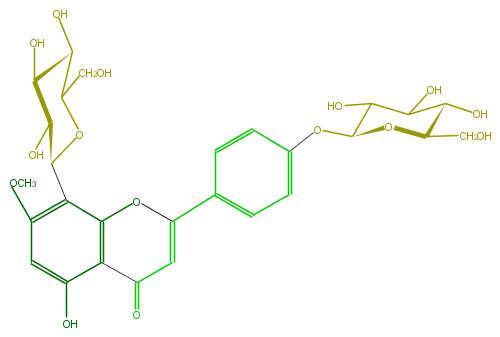
| − | ID FL3FCADS0003 | + | ID FL3FCADS0003 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES C(=O)(c12)C=C(c(c5)ccc(c5)OC(O4)C(O)C(O)C(C4CO)O)Oc1c(C(C(O)3)OC(CO)C(O)C3O)c(cc(O)2)OC | + | SMILES C(=O)(c12)C=C(c(c5)ccc(c5)OC(O4)C(O)C(O)C(C4CO)O)Oc1c(C(C(O)3)OC(CO)C(O)C3O)c(cc(O)2)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.9610 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9610 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2466 -2.3150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5322 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5322 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2466 -0.6651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8177 -2.3150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1033 -1.9025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1033 -1.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8177 -0.6651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8177 -2.9581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1229 2.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9006 1.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5707 0.9149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5619 0.0964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9669 0.6912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3586 1.4335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4489 3.1396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9027 2.5362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9316 0.2831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2466 -3.1396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2303 -0.5327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5227 -0.9674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2755 -0.5327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2755 0.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5227 0.7712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2303 0.3366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8422 0.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9301 1.3007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5438 0.6317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2866 0.8439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0341 0.6317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4205 1.3007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6776 1.0886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2759 1.2721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1242 1.5935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0619 1.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4198 -0.2829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0619 0.8292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9174 1.9480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9998 1.4183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7868 0.6673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7868 -0.3921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
12 13 1 1 0 0 0
13 14 1 1 0 0 0
15 14 1 1 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 12 1 0 0 0 0
12 18 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
6 15 1 0 0 0 0
3 21 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 22 1 0 0 0 0
9 22 1 0 0 0 0
25 28 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
30 28 1 0 0 0 0
38 39 1 0 0 0 0
1 38 1 0 0 0 0
40 41 1 0 0 0 0
17 40 1 0 0 0 0
42 43 1 0 0 0 0
32 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 ^ OCH3
M SBV 1 43 0.4587 -0.7946
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 45
M SMT 2 CH2OH
M SBV 2 45 -0.4412 -0.5145
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 47
M SMT 3 CH2OH
M SBV 3 47 -0.7527 -0.0356
S SKP 5
ID FL3FCADS0003
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES C(=O)(c12)C=C(c(c5)ccc(c5)OC(O4)C(O)C(O)C(C4CO)O)Oc1c(C(C(O)3)OC(CO)C(O)C3O)c(cc(O)2)OC
M END