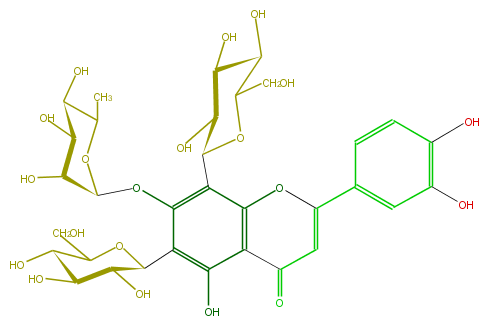
Mol:FL3FACDS0023
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4334 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4334 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4334 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4334 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7189 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7189 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0045 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0045 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0045 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0045 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7189 -0.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7189 -0.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7100 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7100 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4245 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4245 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4245 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4245 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7100 -0.5065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7100 -0.5065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7100 -2.7997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7100 -2.7997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1476 -0.5067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1476 -0.5067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3415 2.1187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3415 2.1187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5909 1.8310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5909 1.8310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5710 0.9209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5710 0.9209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8427 0.1485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8427 0.1485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0800 0.5041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0800 0.5041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1943 1.3355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1943 1.3355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2063 2.9812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2063 2.9812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3792 2.5224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3792 2.5224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2759 0.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2759 0.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7189 -2.9812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7189 -2.9812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2182 -0.4801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2182 -0.4801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9710 -0.9149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9710 -0.9149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7239 -0.4801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7239 -0.4801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7239 0.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7239 0.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9710 0.8238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9710 0.8238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2182 0.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2182 0.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4763 0.8236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4763 0.8236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8383 -1.7760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8383 -1.7760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3617 -2.4052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3617 -2.4052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6753 -2.1383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6753 -2.1383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0128 -2.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0128 -2.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4942 -1.6497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4942 -1.6497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0947 -1.9667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0947 -1.9667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4763 -2.0347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4763 -2.0347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1035 -2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1035 -2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0949 -2.6143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0949 -2.6143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6279 -0.2848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6279 -0.2848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9534 -0.6743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9534 -0.6743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1675 0.0747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1675 0.0747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9534 0.8281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9534 0.8281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6279 1.2176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6279 1.2176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4139 0.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4139 0.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3406 1.8322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3406 1.8322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9534 1.3141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9534 1.3141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0194 0.8979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0194 0.8979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2582 -0.3151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2582 -0.3151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3582 -0.8464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3582 -0.8464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4257 1.6083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4257 1.6083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1068 0.7966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1068 0.7966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7735 -1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7735 -1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8559 -1.9379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8559 -1.9379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 39 1 1 0 0 0 | + | 44 39 1 1 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 42 46 1 0 0 0 0 | + | 42 46 1 0 0 0 0 |
| − | 44 47 1 0 0 0 0 | + | 44 47 1 0 0 0 0 |
| − | 39 48 1 0 0 0 0 | + | 39 48 1 0 0 0 0 |
| − | 40 12 1 0 0 0 0 | + | 40 12 1 0 0 0 0 |
| − | 25 49 1 0 0 0 0 | + | 25 49 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 18 50 1 0 0 0 0 | + | 18 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 35 52 1 0 0 0 0 | + | 35 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 56 -0.6200 -0.2728 | + | M SBV 1 56 -0.6200 -0.2728 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 58 0.6788 -0.5586 | + | M SBV 2 58 0.6788 -0.5586 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACDS0023 | + | ID FL3FACDS0023 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(O)(C(O)1)C(OC(c(c(OC(C(O)6)OC(C(C6O)O)C)4)c(O2)c(c(O)c(C(C(O)5)OC(C(O)C5O)CO)4)C(=O)C=C2c(c3)ccc(c3O)O)C1O)CO | + | SMILES C(O)(C(O)1)C(OC(c(c(OC(C(O)6)OC(C(C6O)O)C)4)c(O2)c(c(O)c(C(C(O)5)OC(C(O)C5O)CO)4)C(=O)C=C2c(c3)ccc(c3O)O)C1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.4334 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4334 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7189 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0045 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0045 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7189 -0.5065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7100 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4245 -1.7440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4245 -0.9190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7100 -0.5065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7100 -2.7997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1476 -0.5067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3415 2.1187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5909 1.8310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5710 0.9209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8427 0.1485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0800 0.5041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1943 1.3355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2063 2.9812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3792 2.5224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2759 0.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7189 -2.9812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2182 -0.4801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9710 -0.9149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7239 -0.4801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7239 0.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9710 0.8238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2182 0.3892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4763 0.8236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8383 -1.7760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3617 -2.4052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6753 -2.1383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0128 -2.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4942 -1.6497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0947 -1.9667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4763 -2.0347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1035 -2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0949 -2.6143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6279 -0.2848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9534 -0.6743 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1675 0.0747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9534 0.8281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6279 1.2176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4139 0.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3406 1.8322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9534 1.3141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0194 0.8979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2582 -0.3151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3582 -0.8464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4257 1.6083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1068 0.7966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7735 -1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8559 -1.9379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
40 39 1 1 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 1 0 0 0
44 39 1 1 0 0 0
43 45 1 0 0 0 0
42 46 1 0 0 0 0
44 47 1 0 0 0 0
39 48 1 0 0 0 0
40 12 1 0 0 0 0
25 49 1 0 0 0 0
50 51 1 0 0 0 0
18 50 1 0 0 0 0
52 53 1 0 0 0 0
35 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 CH2OH
M SBV 1 56 -0.6200 -0.2728
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 ^CH2OH
M SBV 2 58 0.6788 -0.5586
S SKP 5
ID FL3FACDS0023
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(O)(C(O)1)C(OC(c(c(OC(C(O)6)OC(C(C6O)O)C)4)c(O2)c(c(O)c(C(C(O)5)OC(C(O)C5O)CO)4)C(=O)C=C2c(c3)ccc(c3O)O)C1O)CO
M END