Mol:FL3FACCS0059
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.2409 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2409 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5265 0.5662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5265 0.5662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8120 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8120 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8120 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8120 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5265 -1.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5265 -1.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2409 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2409 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0975 0.5662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0975 0.5662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3831 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3831 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3831 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3831 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0975 -1.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0975 -1.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5265 -1.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5265 -1.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0423 0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0423 0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7568 0.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7568 0.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4713 0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4713 0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4713 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4713 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7568 1.8538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7568 1.8538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0423 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0423 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1547 1.8359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1547 1.8359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0975 -1.8538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0975 -1.8538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3074 0.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3074 0.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1328 0.2344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1328 0.2344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4702 -0.3079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4702 -0.3079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0576 -1.0226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0576 -1.0226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2592 -0.8134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2592 -0.8134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4658 -1.0403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4658 -1.0403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8783 -0.3255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8783 -0.3255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6767 -0.5346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6767 -0.5346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8337 0.0512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8337 0.0512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9534 -0.7911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9534 -0.7911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5063 -0.7635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5063 -0.7635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4775 -1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4775 -1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3807 0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3807 0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7926 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7926 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3807 1.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3807 1.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6165 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6165 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0933 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0933 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9183 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9183 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3308 1.1342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3308 1.1342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9183 0.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9183 0.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0933 0.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0933 0.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.1547 1.1342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.1547 1.1342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 25 9 1 0 0 0 0 | + | 25 9 1 0 0 0 0 |
| − | 31 24 1 0 0 0 0 | + | 31 24 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 32 28 1 0 0 0 0 | + | 32 28 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACCS0059 | + | ID FL3FACCS0059 |
| − | KNApSAcK_ID C00014101 | + | KNApSAcK_ID C00014101 |
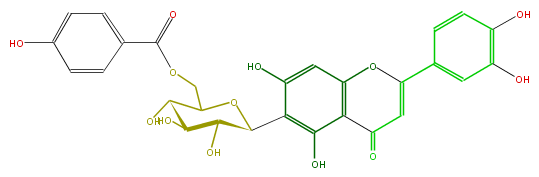
| − | NAME Perfoliatumin A;Isoorientin 6''-O-p-hydroxybenzoate | + | NAME Perfoliatumin A;Isoorientin 6''-O-p-hydroxybenzoate |
| − | CAS_RN 325772-31-4 | + | CAS_RN 325772-31-4 |
| − | FORMULA C28H24O13 | + | FORMULA C28H24O13 |
| − | EXACTMASS 568.121690854 | + | EXACTMASS 568.121690854 |
| − | AVERAGEMASS 568.48236 | + | AVERAGEMASS 568.48236 |
| − | SMILES c(c5O)(ccc(c5)C(O1)=CC(=O)c(c(O)2)c(cc(O)c2C(C4O)OC(C(C(O)4)O)COC(c(c3)ccc(O)c3)=O)1)O | + | SMILES c(c5O)(ccc(c5)C(O1)=CC(=O)c(c(O)2)c(cc(O)c2C(C4O)OC(C(C(O)4)O)COC(c(c3)ccc(O)c3)=O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
3.2409 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5265 0.5662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8120 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8120 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5265 -1.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2409 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0975 0.5662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3831 0.1537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3831 -0.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0975 -1.0838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5265 -1.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0423 0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7568 0.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4713 0.6163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4713 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7568 1.8538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0423 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1547 1.8359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0975 -1.8538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3074 0.5523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1328 0.2344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4702 -0.3079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0576 -1.0226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2592 -0.8134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4658 -1.0403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8783 -0.3255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6767 -0.5346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8337 0.0512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9534 -0.7911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5063 -0.7635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4775 -1.5613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3807 0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7926 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3807 1.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6165 1.1217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0933 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9183 1.8486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3308 1.1342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9183 0.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0933 0.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.1547 1.1342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
3 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
1 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
10 19 1 0 0 0 0
8 20 1 0 0 0 0
14 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
25 9 1 0 0 0 0
31 24 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
38 41 1 0 0 0 0
32 28 1 0 0 0 0
35 36 2 0 0 0 0
35 40 1 0 0 0 0
S SKP 8
ID FL3FACCS0059
KNApSAcK_ID C00014101
NAME Perfoliatumin A;Isoorientin 6''-O-p-hydroxybenzoate
CAS_RN 325772-31-4
FORMULA C28H24O13
EXACTMASS 568.121690854
AVERAGEMASS 568.48236
SMILES c(c5O)(ccc(c5)C(O1)=CC(=O)c(c(O)2)c(cc(O)c2C(C4O)OC(C(C(O)4)O)COC(c(c3)ccc(O)c3)=O)1)O
M END