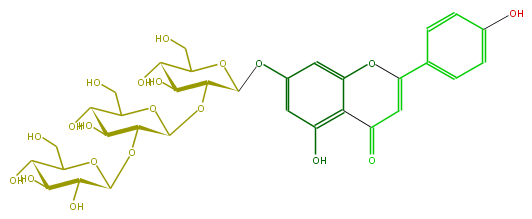
Mol:FL3FAAGS0059
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.6240 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6240 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6240 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6240 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3385 -0.4568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3385 -0.4568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0529 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0529 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0529 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0529 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3385 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3385 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7674 -0.4568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7674 -0.4568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4819 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4819 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4819 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4819 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7674 1.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7674 1.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1963 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1963 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9108 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9108 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6253 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6253 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6253 2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6253 2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9108 2.4307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9108 2.4307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1963 2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1963 2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7674 -1.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7674 -1.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3385 -1.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3385 -1.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0905 1.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0905 1.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3716 2.4490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3716 2.4490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6238 1.1906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6238 1.1906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2112 0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2112 0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4128 0.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4128 0.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6194 0.4582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6194 0.4582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0319 1.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0319 1.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8303 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8303 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9873 1.5497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9873 1.5497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4671 1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4671 1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1070 0.7074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1070 0.7074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6598 0.7350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6598 0.7350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5843 0.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5843 0.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3899 0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3899 0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9772 -0.6629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9772 -0.6629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1789 -0.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1789 -0.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3854 -0.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3854 -0.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7980 0.0341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7980 0.0341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5964 -0.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5964 -0.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7533 0.4109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7533 0.4109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2332 0.6879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2332 0.6879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8731 -0.4314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8731 -0.4314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4259 -0.4039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4259 -0.4039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3504 -1.0937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3504 -1.0937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8884 -1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8884 -1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4757 -2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4757 -2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6774 -1.8091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6774 -1.8091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8839 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8839 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2965 -1.3212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2965 -1.3212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0949 -1.5303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0949 -1.5303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2519 -0.9444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2519 -0.9444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7317 -0.6674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7317 -0.6674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.3716 -1.7867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.3716 -1.7867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9244 -1.7592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9244 -1.7592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8489 -2.4490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8489 -2.4490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 43 51 1 0 0 0 0 | + | 43 51 1 0 0 0 0 |
| − | 44 52 1 0 0 0 0 | + | 44 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 46 42 1 0 0 0 0 | + | 46 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0059 | + | ID FL3FAAGS0059 |
| − | KNApSAcK_ID C00013609 | + | KNApSAcK_ID C00013609 |
| − | NAME Apigenin 7-sophorotrioside;5,7,4'-Trihydroxyflavone 7-sophorotrioside;7-[(O-beta-D-Glucopyranosyl-(1->2)-O-beta-D-glucopyranosyl-(1->2)-b-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one | + | NAME Apigenin 7-sophorotrioside;5,7,4'-Trihydroxyflavone 7-sophorotrioside;7-[(O-beta-D-Glucopyranosyl-(1->2)-O-beta-D-glucopyranosyl-(1->2)-b-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 205326-78-9 | + | CAS_RN 205326-78-9 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES OC(C(CO)6)C(O)C(O)C(O6)OC(C1OC(C2O)C(Oc(c3)cc(c(C5=O)c3OC(=C5)c(c4)ccc(c4)O)O)OC(C2O)CO)C(O)C(C(CO)O1)O | + | SMILES OC(C(CO)6)C(O)C(O)C(O6)OC(C1OC(C2O)C(Oc(c3)cc(c(C5=O)c3OC(=C5)c(c4)ccc(c4)O)O)OC(C2O)CO)C(O)C(C(CO)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
0.6240 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6240 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3385 -0.4568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0529 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0529 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3385 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7674 -0.4568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4819 -0.0443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4819 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7674 1.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1963 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9108 0.7807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6253 1.1932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6253 2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9108 2.4307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1963 2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7674 -1.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3385 -1.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0905 1.1932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.3716 2.4490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6238 1.1906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2112 0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4128 0.6851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6194 0.4582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0319 1.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8303 0.9639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9873 1.5497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4671 1.8268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1070 0.7074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6598 0.7350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5843 0.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3899 0.0518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9772 -0.6629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1789 -0.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3854 -0.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7980 0.0341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5964 -0.1750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7533 0.4109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2332 0.6879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8731 -0.4314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4259 -0.4039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3504 -1.0937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.8884 -1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4757 -2.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6774 -1.8091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8839 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2965 -1.3212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0949 -1.5303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2519 -0.9444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7317 -0.6674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.3716 -1.7867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9244 -1.7592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8489 -2.4490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
14 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
21 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
35 31 1 0 0 0 0
24 19 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
43 51 1 0 0 0 0
44 52 1 0 0 0 0
45 53 1 0 0 0 0
46 42 1 0 0 0 0
S SKP 8
ID FL3FAAGS0059
KNApSAcK_ID C00013609
NAME Apigenin 7-sophorotrioside;5,7,4'-Trihydroxyflavone 7-sophorotrioside;7-[(O-beta-D-Glucopyranosyl-(1->2)-O-beta-D-glucopyranosyl-(1->2)-b-D-glucopyranosyl)oxy]-5-hydroxy-2-(4-hydroxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 205326-78-9
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES OC(C(CO)6)C(O)C(O)C(O6)OC(C1OC(C2O)C(Oc(c3)cc(c(C5=O)c3OC(=C5)c(c4)ccc(c4)O)O)OC(C2O)CO)C(O)C(C(CO)O1)O
M END