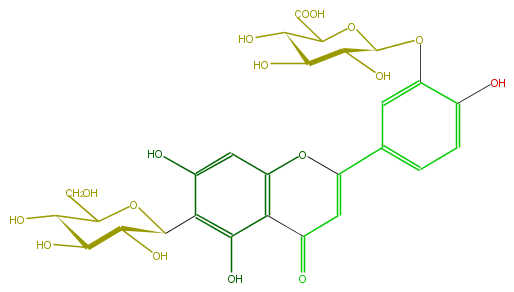
Mol:FL3FAADS0026
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1949 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1949 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1949 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1949 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4805 -2.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4805 -2.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2340 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2340 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2340 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2340 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4805 -0.6272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4805 -0.6272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9484 -2.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9484 -2.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6630 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6630 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6630 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6630 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9484 -0.6272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9484 -0.6272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9484 -3.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9484 -3.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4805 -3.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4805 -3.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5360 -0.4949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5360 -0.4949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2888 -0.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2888 -0.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0416 -0.4949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0416 -0.4949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0416 0.3744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0416 0.3744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2888 0.8090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2888 0.8090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5360 0.3744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5360 0.3744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7941 0.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7941 0.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9982 -1.8501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9982 -1.8501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3580 -2.5079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3580 -2.5079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6959 -2.1105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6959 -2.1105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7952 -2.2165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7952 -2.2165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4309 -1.6338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4309 -1.6338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1461 -1.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1461 -1.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6858 -1.9244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6858 -1.9244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1564 -2.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1564 -2.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9299 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9299 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9672 -2.6557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9672 -2.6557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2888 1.6777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2888 1.6777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5853 1.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5853 1.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0620 1.1733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0620 1.1733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7484 1.4402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7484 1.4402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4108 1.4474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4108 1.4474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9295 1.9288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9295 1.9288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3290 1.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3290 1.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1142 1.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1142 1.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1784 1.1758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1784 1.1758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4938 0.9436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4938 0.9436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8338 -1.3893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8338 -1.3893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7941 -0.9415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7941 -0.9415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7650 2.1835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7650 2.1835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4822 2.4890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4822 2.4890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6533 3.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6533 3.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 17 30 1 0 0 0 0 | + | 17 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.6877 -0.5888 | + | M SBV 1 45 0.6877 -0.5888 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 42 43 44 | + | M SAL 2 3 42 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 ^ COOH | + | M SMT 2 ^ COOH |
| − | M SBV 2 48 0.5640 -0.5716 | + | M SBV 2 48 0.5640 -0.5716 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAADS0026 | + | ID FL3FAADS0026 |
| − | FORMULA C27H28O17 | + | FORMULA C27H28O17 |
| − | EXACTMASS 624.1326494699999 | + | EXACTMASS 624.1326494699999 |
| − | AVERAGEMASS 624.50102 | + | AVERAGEMASS 624.50102 |
| − | SMILES OCC(O1)C(O)C(O)C(C(c(c2O)c(cc(O3)c(C(C=C3c(c4)cc(OC(C5O)OC(C(O)C5O)C(O)=O)c(c4)O)=O)2)O)1)O | + | SMILES OCC(O1)C(O)C(O)C(C(c(c2O)c(cc(O3)c(C(C=C3c(c4)cc(OC(C5O)OC(C(O)C5O)C(O)=O)c(c4)O)=O)2)O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.1949 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1949 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4805 -2.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2340 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2340 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4805 -0.6272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9484 -2.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6630 -1.8647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6630 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9484 -0.6272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9484 -3.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4805 -3.1018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5360 -0.4949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2888 -0.9297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0416 -0.4949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0416 0.3744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2888 0.8090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5360 0.3744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7941 0.8087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9982 -1.8501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3580 -2.5079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6959 -2.1105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7952 -2.2165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4309 -1.6338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1461 -1.9781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6858 -1.9244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1564 -2.4254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9299 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9672 -2.6557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2888 1.6777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5853 1.8025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0620 1.1733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7484 1.4402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4108 1.4474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9295 1.9288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3290 1.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1142 1.6870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1784 1.1758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4938 0.9436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8338 -1.3893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7941 -0.9415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7650 2.1835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4822 2.4890 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6533 3.1110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
17 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 30 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
36 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.6877 -0.5888
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 42 43 44
M SBL 2 1 48
M SMT 2 ^ COOH
M SBV 2 48 0.5640 -0.5716
S SKP 5
ID FL3FAADS0026
FORMULA C27H28O17
EXACTMASS 624.1326494699999
AVERAGEMASS 624.50102
SMILES OCC(O1)C(O)C(O)C(C(c(c2O)c(cc(O3)c(C(C=C3c(c4)cc(OC(C5O)OC(C(O)C5O)C(O)=O)c(c4)O)=O)2)O)1)O
M END