Mol:FL2FCBGM0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.5250 0.7861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5250 0.7861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5250 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5250 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9687 -0.1774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9687 -0.1774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4124 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4124 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4124 0.7861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4124 0.7861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9687 1.1073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9687 1.1073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1439 -0.1774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1439 -0.1774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7002 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7002 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7002 0.7861 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.7002 0.7861 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.1439 1.1073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1439 1.1073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2563 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2563 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8232 0.7799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8232 0.7799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3902 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3902 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3902 1.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3902 1.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8232 2.0892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8232 2.0892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2563 1.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2563 1.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1439 -0.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1439 -0.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9687 -0.8195 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9687 -0.8195 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1113 -0.1947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1113 -0.1947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9687 1.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9687 1.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4638 -1.3836 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.4638 -1.3836 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.9170 -1.9303 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.9170 -1.9303 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2452 -1.5475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.2452 -1.5475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.4721 -1.5475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.4721 -1.5475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.0188 -1.0006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0188 -1.0006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6907 -1.3836 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.6907 -1.3836 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.9893 -1.9907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9893 -1.9907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5100 -2.0892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5100 -2.0892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4604 -1.3016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4604 -1.3016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9570 2.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9570 2.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6715 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6715 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8823 1.4049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8823 1.4049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3823 2.2709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3823 2.2709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7753 -0.9641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7753 -0.9641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5230 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5230 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 2 19 1 0 0 0 0 | + | 2 19 1 0 0 0 0 |
| − | 6 20 1 0 0 0 0 | + | 6 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 14 30 1 0 0 0 0 | + | 14 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 1 32 1 0 0 0 0 | + | 1 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 26 34 1 0 0 0 0 | + | 26 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 34 35 | + | M SAL 3 2 34 35 |
| − | M SBL 3 1 37 | + | M SBL 3 1 37 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 37 -2.8469 -0.9852 | + | M SVB 3 37 -2.8469 -0.9852 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 35 | + | M SBL 2 1 35 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 35 -1.8823 1.4049 | + | M SVB 2 35 -1.8823 1.4049 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 31 | + | M SAL 1 2 30 31 |
| − | M SBL 1 1 33 | + | M SBL 1 1 33 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 33 2.957 2.0891 | + | M SVB 1 33 2.957 2.0891 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FCBGM0001 | + | ID FL2FCBGM0001 |
| − | KNApSAcK_ID C00008264 | + | KNApSAcK_ID C00008264 |
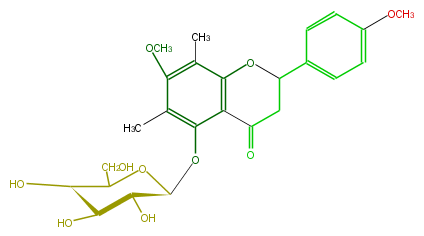
| − | NAME 5-Hydroxy-7,4'-dimethoxy-6,8-di-C-prenylflavanone 5-O-galactoside | + | NAME 5-Hydroxy-7,4'-dimethoxy-6,8-di-C-prenylflavanone 5-O-galactoside |
| − | CAS_RN 85687-90-7 | + | CAS_RN 85687-90-7 |
| − | FORMULA C25H30O10 | + | FORMULA C25H30O10 |
| − | EXACTMASS 490.18389718 | + | EXACTMASS 490.18389718 |
| − | AVERAGEMASS 490.49969999999996 | + | AVERAGEMASS 490.49969999999996 |
| − | SMILES O[C@H]([C@@H]1Oc(c4C)c(c3c(c4OC)C)C(=O)CC(O3)c(c2)ccc(OC)c2)[C@H]([C@@H](O)C(CO)O1)O | + | SMILES O[C@H]([C@@H]1Oc(c4C)c(c3c(c4OC)C)C(=O)CC(O3)c(c2)ccc(OC)c2)[C@H]([C@@H](O)C(CO)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-1.5250 0.7861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5250 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9687 -0.1774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4124 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4124 0.7861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9687 1.1073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1439 -0.1774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7002 0.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7002 0.7861 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.1439 1.1073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2563 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8232 0.7799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3902 1.1072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3902 1.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8232 2.0892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2563 1.7619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1439 -0.7212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9687 -0.8195 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1113 -0.1947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9687 1.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4638 -1.3836 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.9170 -1.9303 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2452 -1.5475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.4721 -1.5475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.0188 -1.0006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6907 -1.3836 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.9893 -1.9907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5100 -2.0892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4604 -1.3016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9570 2.0891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6715 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8823 1.4049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3823 2.2709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7753 -0.9641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5230 -0.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
3 18 1 0 0 0 0
2 19 1 0 0 0 0
6 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
18 24 1 0 0 0 0
23 27 1 0 0 0 0
22 28 1 0 0 0 0
21 29 1 0 0 0 0
14 30 1 0 0 0 0
30 31 1 0 0 0 0
1 32 1 0 0 0 0
32 33 1 0 0 0 0
26 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 34 35
M SBL 3 1 37
M SMT 3 CH2OH
M SVB 3 37 -2.8469 -0.9852
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 35
M SMT 2 OCH3
M SVB 2 35 -1.8823 1.4049
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 31
M SBL 1 1 33
M SMT 1 OCH3
M SVB 1 33 2.957 2.0891
S SKP 8
ID FL2FCBGM0001
KNApSAcK_ID C00008264
NAME 5-Hydroxy-7,4'-dimethoxy-6,8-di-C-prenylflavanone 5-O-galactoside
CAS_RN 85687-90-7
FORMULA C25H30O10
EXACTMASS 490.18389718
AVERAGEMASS 490.49969999999996
SMILES O[C@H]([C@@H]1Oc(c4C)c(c3c(c4OC)C)C(=O)CC(O3)c(c2)ccc(OC)c2)[C@H]([C@@H](O)C(CO)O1)O
M END