Mol:FL2FAAGS0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.5041 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5041 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5041 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5041 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0604 -1.2232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0604 -1.2232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6167 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6167 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6167 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6167 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0604 0.0615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0604 0.0615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1730 -1.2232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1730 -1.2232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7293 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7293 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7293 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7293 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1730 0.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1730 0.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2854 0.0614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2854 0.0614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8524 -0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8524 -0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4194 0.0614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4194 0.0614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4194 0.7161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4194 0.7161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8524 1.0434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8524 1.0434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2854 0.7161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2854 0.7161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1730 -1.7670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1730 -1.7670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0522 0.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0522 0.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9863 1.0434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9863 1.0434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0604 -1.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0604 -1.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1162 -0.0719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1162 -0.0719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7480 -0.5578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7480 -0.5578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2179 -0.3517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2179 -0.3517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7063 -0.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7063 -0.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0781 0.0256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0781 0.0256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5419 -0.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5419 -0.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6259 -0.3661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6259 -0.3661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3220 -0.5858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3220 -0.5858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9141 -0.8616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9141 -0.8616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9679 0.5187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9679 0.5187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9679 1.1271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9679 1.1271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5042 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5042 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5371 0.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5371 0.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8957 0.5556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8957 0.5556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4508 0.0481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4508 0.0481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4178 0.7222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4178 0.7222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0592 0.9321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0592 0.9321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0275 0.0638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0275 0.0638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9863 1.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9863 1.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2583 1.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2583 1.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8298 1.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8298 1.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0329 1.3294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0329 1.3294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -6.3203 5.6476 | + | M SBV 1 45 -6.3203 5.6476 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FAAGS0016 | + | ID FL2FAAGS0016 |
| − | KNApSAcK_ID C00008216 | + | KNApSAcK_ID C00008216 |
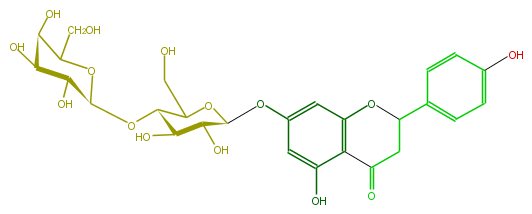
| − | NAME 5,7,4'-Trihydroxyflavanone 7-O-galactosylglucoside | + | NAME 5,7,4'-Trihydroxyflavanone 7-O-galactosylglucoside |
| − | CAS_RN 81244-23-7 | + | CAS_RN 81244-23-7 |
| − | FORMULA C27H32O15 | + | FORMULA C27H32O15 |
| − | EXACTMASS 596.174120354 | + | EXACTMASS 596.174120354 |
| − | AVERAGEMASS 596.5339799999999 | + | AVERAGEMASS 596.5339799999999 |
| − | SMILES C(C4c(c5)ccc(c5)O)C(=O)c(c(O4)3)c(O)cc(c3)OC(O1)C(C(O)C(OC(O2)C(O)C(O)C(O)C2CO)C1CO)O | + | SMILES C(C4c(c5)ccc(c5)O)C(=O)c(c(O4)3)c(O)cc(c3)OC(O1)C(C(O)C(OC(O2)C(O)C(O)C(O)C2CO)C1CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.5041 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5041 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0604 -1.2232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6167 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6167 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0604 0.0615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1730 -1.2232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7293 -0.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7293 -0.2597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1730 0.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2854 0.0614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8524 -0.2660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4194 0.0614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4194 0.7161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8524 1.0434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2854 0.7161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1730 -1.7670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0522 0.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9863 1.0434 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0604 -1.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1162 -0.0719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7480 -0.5578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2179 -0.3517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7063 -0.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0781 0.0256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5419 -0.2191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6259 -0.3661 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3220 -0.5858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9141 -0.8616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9679 0.5187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9679 1.1271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5042 1.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5371 0.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8957 0.5556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4508 0.0481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4178 0.7222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0592 0.9321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0275 0.0638 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9863 1.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2583 1.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8298 1.5429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0329 1.3294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
18 1 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
26 30 1 0 0 0 0
30 31 1 0 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
27 35 1 0 0 0 0
34 38 1 0 0 0 0
33 39 1 0 0 0 0
32 40 1 0 0 0 0
37 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -6.3203 5.6476
S SKP 8
ID FL2FAAGS0016
KNApSAcK_ID C00008216
NAME 5,7,4'-Trihydroxyflavanone 7-O-galactosylglucoside
CAS_RN 81244-23-7
FORMULA C27H32O15
EXACTMASS 596.174120354
AVERAGEMASS 596.5339799999999
SMILES C(C4c(c5)ccc(c5)O)C(=O)c(c(O4)3)c(O)cc(c3)OC(O1)C(C(O)C(OC(O2)C(O)C(O)C(O)C2CO)C1CO)O
M END