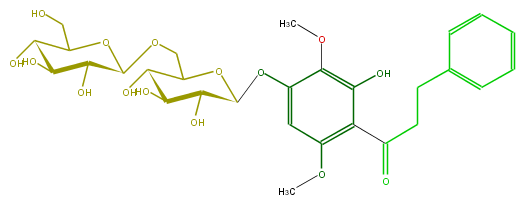
Mol:FL1DE9GS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 47 0 0 0 0 0 0 0 0999 V2000 | + | 44 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.6030 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6030 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6030 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6030 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3174 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3174 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0319 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0319 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0319 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0319 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3174 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3174 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7464 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7464 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4609 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4609 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4609 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4609 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1753 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1753 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8898 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8898 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6043 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6043 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6043 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6043 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8898 1.9457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8898 1.9457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1753 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1753 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7464 -1.7668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7464 -1.7668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6224 0.6367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6224 0.6367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0554 0.6758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0554 0.6758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3174 1.4012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3174 1.4012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6776 1.7707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6776 1.7707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3174 -1.6185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3174 -1.6185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6588 -1.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6588 -1.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5801 0.7026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5801 0.7026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1674 -0.0121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1674 -0.0121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3691 0.1970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3691 0.1970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5756 -0.0298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5756 -0.0298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9882 0.6849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9882 0.6849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7866 0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7866 0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9436 1.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9436 1.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4234 1.3387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4234 1.3387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0633 0.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0633 0.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6161 0.2469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6161 0.2469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5406 -0.4429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5406 -0.4429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1211 1.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1211 1.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7084 0.6479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7084 0.6479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9101 0.8570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9101 0.8570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1166 0.6302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1166 0.6302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5292 1.3449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5292 1.3449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3276 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3276 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4846 1.7217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4846 1.7217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9644 1.9987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9644 1.9987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6043 0.8794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6043 0.8794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1571 0.9069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1571 0.9069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0816 0.2171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0816 0.2171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 5 17 1 0 0 0 0 | + | 5 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 6 19 1 0 0 0 0 | + | 6 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 18 1 0 0 0 0 | + | 26 18 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 37 30 1 0 0 0 0 | + | 37 30 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1DE9GS0001 | + | ID FL1DE9GS0001 |
| − | KNApSAcK_ID C00014627 | + | KNApSAcK_ID C00014627 |
| − | NAME Salicifolioside A;2',4'-Dihydroxy-3',6'-dimethoxydihydrochalcone 4'-glucosyl-(1'''->6'')-glucoside;2',4'-Dihydroxy-3',6'-dimethoxydihydrochalcone 4'-gentiobioside | + | NAME Salicifolioside A;2',4'-Dihydroxy-3',6'-dimethoxydihydrochalcone 4'-glucosyl-(1'''->6'')-glucoside;2',4'-Dihydroxy-3',6'-dimethoxydihydrochalcone 4'-gentiobioside |
| − | CAS_RN 243130-37-2 | + | CAS_RN 243130-37-2 |
| − | FORMULA C29H38O15 | + | FORMULA C29H38O15 |
| − | EXACTMASS 626.221070546 | + | EXACTMASS 626.221070546 |
| − | AVERAGEMASS 626.60302 | + | AVERAGEMASS 626.60302 |
| − | SMILES c(c3C(=O)CCc(c4)cccc4)(OC)cc(c(c3O)OC)OC(O1)C(C(O)C(C1COC(C(O)2)OC(CO)C(O)C2O)O)O | + | SMILES c(c3C(=O)CCc(c4)cccc4)(OC)cc(c(c3O)OC)OC(O1)C(C(O)C(C1COC(C(O)2)OC(CO)C(O)C2O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 47 0 0 0 0 0 0 0 0999 V2000
0.6030 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6030 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3174 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0319 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0319 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3174 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7464 -0.9418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4609 -0.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4609 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1753 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8898 0.2957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6043 0.7082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6043 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8898 1.9457 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1753 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7464 -1.7668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6224 0.6367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0554 0.6758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3174 1.4012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6776 1.7707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3174 -1.6185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6588 -1.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5801 0.7026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1674 -0.0121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3691 0.1970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5756 -0.0298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9882 0.6849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7866 0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9436 1.0617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4234 1.3387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0633 0.2194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6161 0.2469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5406 -0.4429 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1211 1.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7084 0.6479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9101 0.8570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1166 0.6302 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5292 1.3449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3276 1.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4846 1.7217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9644 1.9987 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.6043 0.8794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1571 0.9069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0816 0.2171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
5 17 1 0 0 0 0
1 18 1 0 0 0 0
6 19 1 0 0 0 0
19 20 1 0 0 0 0
3 21 1 0 0 0 0
21 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 18 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
35 43 1 0 0 0 0
36 44 1 0 0 0 0
37 30 1 0 0 0 0
S SKP 8
ID FL1DE9GS0001
KNApSAcK_ID C00014627
NAME Salicifolioside A;2',4'-Dihydroxy-3',6'-dimethoxydihydrochalcone 4'-glucosyl-(1'''->6'')-glucoside;2',4'-Dihydroxy-3',6'-dimethoxydihydrochalcone 4'-gentiobioside
CAS_RN 243130-37-2
FORMULA C29H38O15
EXACTMASS 626.221070546
AVERAGEMASS 626.60302
SMILES c(c3C(=O)CCc(c4)cccc4)(OC)cc(c(c3O)OC)OC(O1)C(C(O)C(C1COC(C(O)2)OC(CO)C(O)C2O)O)O
M END