Mol:FL1AACGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.0120 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0120 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0120 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0120 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5312 1.1063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5312 1.1063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0505 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0505 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0505 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0505 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5312 -0.0928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5312 -0.0928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5582 0.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5582 0.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9106 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9106 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5582 0.9918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5582 0.9918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5697 1.1063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5697 1.1063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3523 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3523 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2362 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2362 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5365 -0.0134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5365 -0.0134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1370 -0.0134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1370 -0.0134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4373 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4373 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1370 1.0269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1370 1.0269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5365 1.0269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5365 1.0269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0368 0.5068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0368 0.5068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7025 -0.5169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7025 -0.5169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5230 1.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5230 1.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1518 0.5597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1518 0.5597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6173 0.7675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6173 0.7675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1016 0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1016 0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4763 1.1479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4763 1.1479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9439 0.9012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9439 0.9012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0368 0.7530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0368 0.7530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7304 0.5315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7304 0.5315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3111 0.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3111 0.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4368 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4368 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5313 -0.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5313 -0.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5330 -0.8891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5330 -0.8891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1618 -1.3791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1618 -1.3791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6273 -1.1712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6273 -1.1712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1116 -1.1657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1116 -1.1657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4863 -0.7908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4863 -0.7908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9539 -1.0376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9539 -1.0376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0468 -1.1858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0468 -1.1858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7404 -1.4073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7404 -1.4073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3211 -1.6853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3211 -1.6853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3424 1.6853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3424 1.6853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5455 1.4718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5455 1.4718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3524 -0.2534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3524 -0.2534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5555 -0.4669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5555 -0.4669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 1 7 1 0 0 0 0 | + | 1 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 2 1 0 0 0 0 | + | 9 2 1 0 0 0 0 |
| − | 4 10 1 0 0 0 0 | + | 4 10 1 0 0 0 0 |
| − | 8 11 2 0 0 0 0 | + | 8 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 7 19 2 0 0 0 0 | + | 7 19 2 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 10 1 0 0 0 0 | + | 23 10 1 0 0 0 0 |
| − | 16 29 1 0 0 0 0 | + | 16 29 1 0 0 0 0 |
| − | 6 30 1 0 0 0 0 | + | 6 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 44 -6.9857 6.6630 | + | M SBV 1 44 -6.9857 6.6630 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 46 -6.9857 6.6630 | + | M SBV 2 46 -6.9857 6.6630 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1AACGS0005 | + | ID FL1AACGS0005 |
| − | KNApSAcK_ID C00008049 | + | KNApSAcK_ID C00008049 |
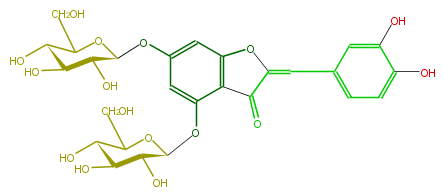
| − | NAME Aureusidin 4,6-diglucoside | + | NAME Aureusidin 4,6-diglucoside |
| − | CAS_RN 89648-26-0 | + | CAS_RN 89648-26-0 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(c(C=c(o5)c(c(c45)c(cc(c4)OC(C3O)OC(CO)C(C(O)3)O)OC(C2O)OC(CO)C(C2O)O)=O)1)c(c(O)cc1)O | + | SMILES c(c(C=c(o5)c(c(c45)c(cc(c4)OC(C3O)OC(CO)C(C(O)3)O)OC(C2O)OC(CO)C(C2O)O)=O)1)c(c(O)cc1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.0120 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0120 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5312 1.1063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0505 0.8065 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0505 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5312 -0.0928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5582 0.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9106 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5582 0.9918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5697 1.1063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3523 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2362 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5365 -0.0134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1370 -0.0134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4373 0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1370 1.0269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5365 1.0269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0368 0.5068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7025 -0.5169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5230 1.0496 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1518 0.5597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6173 0.7675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1016 0.7731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4763 1.1479 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9439 0.9012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0368 0.7530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7304 0.5315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3111 0.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4368 1.5460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5313 -0.6923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5330 -0.8891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1618 -1.3791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6273 -1.1712 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1116 -1.1657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4863 -0.7908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9539 -1.0376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0468 -1.1858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7404 -1.4073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3211 -1.6853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3424 1.6853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5455 1.4718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3524 -0.2534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5555 -0.4669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
1 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 2 1 0 0 0 0
4 10 1 0 0 0 0
8 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
7 19 2 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 10 1 0 0 0 0
16 29 1 0 0 0 0
6 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
30 34 1 0 0 0 0
25 40 1 0 0 0 0
40 41 1 0 0 0 0
36 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 ^CH2OH
M SBV 1 44 -6.9857 6.6630
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 ^CH2OH
M SBV 2 46 -6.9857 6.6630
S SKP 8
ID FL1AACGS0005
KNApSAcK_ID C00008049
NAME Aureusidin 4,6-diglucoside
CAS_RN 89648-26-0
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(c(C=c(o5)c(c(c45)c(cc(c4)OC(C3O)OC(CO)C(C(O)3)O)OC(C2O)OC(CO)C(C2O)O)=O)1)c(c(O)cc1)O
M END