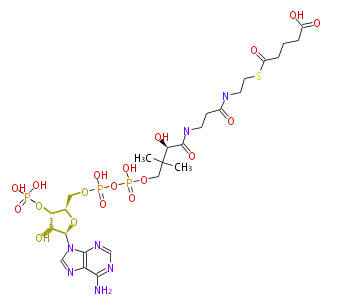
Mol:BMFYS5CAa017
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 58 0 0 1 0 0 0 0 0999 V2000 | + | 56 58 0 0 1 0 0 0 0 0999 V2000 |
| − | 2.9781 -4.3701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9781 -4.3701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6473 -3.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6473 -3.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6254 -3.8348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6254 -3.8348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2946 -3.0917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2946 -3.0917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2727 -3.2996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2727 -3.2996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2872 -5.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2872 -5.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 -4.1621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 -4.1621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5817 -4.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5817 -4.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.6298 -2.2697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.6298 -2.2697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.7638 -1.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.7638 -1.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.7638 -0.7697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.7638 -0.7697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.6298 -0.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.6298 -0.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.4958 -0.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.4958 -0.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.4958 -1.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.4958 -1.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 25.2390 -0.1005 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 25.2390 -0.1005 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.8322 0.8130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.8322 0.8130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.8377 0.7085 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.8377 0.7085 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 25.3619 -2.2697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 25.3619 -2.2697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.1686 1.4516 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 23.1686 1.4516 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 23.3765 2.4298 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 23.3765 2.4298 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 22.5105 2.9298 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 22.5105 2.9298 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 21.7673 2.2607 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 21.7673 2.2607 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 20.7892 2.4686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.7892 2.4686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.2901 2.8365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.2901 2.8365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.4060 3.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.4060 3.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.1741 1.3471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.1741 1.3471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 20.1201 1.7254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.1201 1.7254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.2150 4.5121 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.2150 4.5121 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.8028 3.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.8028 3.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.6272 5.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.6272 5.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.0240 5.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.0240 5.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.1419 1.9333 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.1419 1.9333 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.9340 0.9552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.9340 0.9552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.3498 2.9115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.3498 2.9115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.1638 2.1412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.1638 2.1412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.4946 1.3981 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.4946 1.3981 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.2378 0.7290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.2378 0.7290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.7515 2.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.7515 2.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.8255 0.6550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.8255 0.6550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.8474 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.8474 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.1782 0.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.1782 0.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.9214 -0.5494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.9214 -0.5494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.4351 0.7889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.4351 0.7889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.5091 -0.6234 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 14.5091 -0.6234 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 13.5309 -0.4155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.5309 -0.4155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.8618 -1.1587 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.8618 -1.1587 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.8837 -0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.8837 -0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.2145 -1.6939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.2145 -1.6939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.2364 -1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.2364 -1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.5673 -2.2291 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.5673 -2.2291 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.5891 -2.0212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.5891 -2.0212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.9200 -2.7644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.9200 -2.7644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.9418 -2.5564 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.9418 -2.5564 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.8181 -1.5745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.8181 -1.5745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 13.2219 0.5355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2219 0.5355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9274 -0.5349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9274 -0.5349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9 14 2 0 0 0 0 | + | 9 14 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 13 12 2 0 0 0 0 | + | 13 12 2 0 0 0 0 |
| − | 17 16 1 0 0 0 0 | + | 17 16 1 0 0 0 0 |
| − | 16 15 2 0 0 0 0 | + | 16 15 2 0 0 0 0 |
| − | 15 13 1 0 0 0 0 | + | 15 13 1 0 0 0 0 |
| − | 12 17 1 0 0 0 0 | + | 12 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 22 26 1 6 0 0 0 | + | 22 26 1 6 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 19 26 1 6 0 0 0 | + | 19 26 1 6 0 0 0 |
| − | 21 20 1 0 0 0 0 | + | 21 20 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 19 17 1 0 0 0 0 | + | 19 17 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 35 1 0 0 0 0 | + | 32 35 1 0 0 0 0 |
| − | 39 36 1 0 0 0 0 | + | 39 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 36 35 1 0 0 0 0 | + | 36 35 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 44 1 0 0 0 0 | + | 41 44 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 44 54 1 1 0 0 0 | + | 44 54 1 1 0 0 0 |
| − | 45 55 2 0 0 0 0 | + | 45 55 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 49 56 2 0 0 0 0 | + | 49 56 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 20 24 1 1 0 0 0 | + | 20 24 1 1 0 0 0 |
| − | 21 25 1 1 0 0 0 | + | 21 25 1 1 0 0 0 |
| − | 29 28 1 0 0 0 0 | + | 29 28 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 28 30 2 0 0 0 0 | + | 28 30 2 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 53 5 1 0 0 0 0 | + | 53 5 1 0 0 0 0 |
| − | 5 4 1 0 0 0 0 | + | 5 4 1 0 0 0 0 |
| − | 5 8 2 0 0 0 0 | + | 5 8 2 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 1 6 1 0 0 0 0 | + | 1 6 1 0 0 0 0 |
| − | 1 7 2 0 0 0 0 | + | 1 7 2 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMFYS5CAa017 | + | ID BMFYS5CAa017 |
| − | NAME Glutaryl-CoA | + | NAME Glutaryl-CoA |
| − | FORMULA C26H42N7O19P3S | + | FORMULA C26H42N7O19P3S |
| − | EXACTMASS 881.1469 | + | EXACTMASS 881.1469 |
| − | AVERAGEMASS 881.6347 | + | AVERAGEMASS 881.6347 |
| − | SMILES C([C@H](C(NCCC(NCCSC(CCCC(O)=O)=O)=O)=O)O)(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)3)O[C@H]([C@@H]3O)n(c21)cnc(c(N)ncn2)1)(O)=O)(O)=O | + | SMILES C([C@H](C(NCCC(NCCSC(CCCC(O)=O)=O)=O)=O)O)(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)3)O[C@H]([C@@H]3O)n(c21)cnc(c(N)ncn2)1)(O)=O)(O)=O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00527 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00527 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 58 0 0 1 0 0 0 0 0999 V2000
2.9781 -4.3701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6473 -3.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6254 -3.8348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2946 -3.0917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2727 -3.2996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2872 -5.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 -4.1621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5817 -4.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
23.6298 -2.2697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.7638 -1.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
22.7638 -0.7697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
23.6298 -0.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.4958 -0.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.4958 -1.7697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
25.2390 -0.1005 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
24.8322 0.8130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
23.8377 0.7085 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
25.3619 -2.2697 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
23.1686 1.4516 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
23.3765 2.4298 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
22.5105 2.9298 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
21.7673 2.2607 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
20.7892 2.4686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.2901 2.8365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.4060 3.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.1741 1.3471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
20.1201 1.7254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
23.2150 4.5121 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
23.8028 3.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.6272 5.3211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
24.0240 5.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.1419 1.9333 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
18.9340 0.9552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.3498 2.9115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
18.1638 2.1412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17.4946 1.3981 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
18.2378 0.7290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.7515 2.0672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.8255 0.6550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.8474 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
15.1782 0.1197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
15.9214 -0.5494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.4351 0.7889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.5091 -0.6234 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
13.5309 -0.4155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.8618 -1.1587 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
11.8837 -0.9507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
11.2145 -1.6939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.2364 -1.4860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.5673 -2.2291 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
8.5891 -2.0212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.9200 -2.7644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9418 -2.5564 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
14.8181 -1.5745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
13.2219 0.5355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.9274 -0.5349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9 14 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
13 14 1 0 0 0 0
13 12 2 0 0 0 0
17 16 1 0 0 0 0
16 15 2 0 0 0 0
15 13 1 0 0 0 0
12 17 1 0 0 0 0
14 18 1 0 0 0 0
22 26 1 6 0 0 0
21 22 1 0 0 0 0
19 26 1 6 0 0 0
21 20 1 0 0 0 0
19 20 1 0 0 0 0
22 23 1 0 0 0 0
23 27 1 0 0 0 0
19 17 1 0 0 0 0
27 32 1 0 0 0 0
32 34 1 0 0 0 0
32 33 2 0 0 0 0
32 35 1 0 0 0 0
39 36 1 0 0 0 0
36 37 2 0 0 0 0
36 38 1 0 0 0 0
39 40 1 0 0 0 0
36 35 1 0 0 0 0
40 41 1 0 0 0 0
41 44 1 0 0 0 0
41 42 1 0 0 0 0
41 43 1 0 0 0 0
44 45 1 0 0 0 0
44 54 1 1 0 0 0
45 55 2 0 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
49 56 2 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
52 53 1 0 0 0 0
20 24 1 1 0 0 0
21 25 1 1 0 0 0
29 28 1 0 0 0 0
28 31 1 0 0 0 0
28 30 2 0 0 0 0
25 28 1 0 0 0 0
53 5 1 0 0 0 0
5 4 1 0 0 0 0
5 8 2 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
1 6 1 0 0 0 0
1 7 2 0 0 0 0
2 1 1 0 0 0 0
S SKP 7
ID BMFYS5CAa017
NAME Glutaryl-CoA
FORMULA C26H42N7O19P3S
EXACTMASS 881.1469
AVERAGEMASS 881.6347
SMILES C([C@H](C(NCCC(NCCSC(CCCC(O)=O)=O)=O)=O)O)(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)3)O[C@H]([C@@H]3O)n(c21)cnc(c(N)ncn2)1)(O)=O)(O)=O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00527
M END