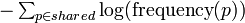Template:MassBank/Matrix
From Metabolomics.JP
(Difference between revisions)
m |
|||
| (68 intermediate revisions by one user not shown) | |||
| Line 1: | Line 1: | ||
{{MassBank/Header}} | {{MassBank/Header}} | ||
| − | + | {{#def:MASSBANKID|{{#substring:{{PAGENAME}}|0|{{#expr:{{#length:{{PAGENAME}}}}-1}}}}}} | |
| + | {{#def:dataline|{{#SearchLine:{{PAGENAME}}|Index|MassBank/Molecules}}}} | ||
<!---化合物組成式の正規表現--------->{{#def:FORMULA_PAT|(C?[1-9]?[0-9]?)(H?[1-9]?[0-9]?)(C?l?[2-9]?)(F?[2-9]?)(I?[2-9]?)(N?[1-9]?[0-9]?)(O?[1-9]?[0-9]?)(P?[2-9]?)(S?[2-9]?)}} | <!---化合物組成式の正規表現--------->{{#def:FORMULA_PAT|(C?[1-9]?[0-9]?)(H?[1-9]?[0-9]?)(C?l?[2-9]?)(F?[2-9]?)(I?[2-9]?)(N?[1-9]?[0-9]?)(O?[1-9]?[0-9]?)(P?[2-9]?)(S?[2-9]?)}} | ||
| − | <!---化合物組成式に出てくる文字列--->{{#def:FORMULA_CHAR|CHFINOPSl0-9}} | + | <!---化合物組成式に出てくる文字列--->{{#def:FORMULA_CHAR|CHFINOPSl0-9%*}} |
<!---化合物組成式に使われる原子----->{{#def:ATOM|{"C", "H", "Cl", "F", "I", "N", "O", "P", "S"} }} | <!---化合物組成式に使われる原子----->{{#def:ATOM|{"C", "H", "Cl", "F", "I", "N", "O", "P", "S"} }} | ||
<!---各原子の質量(上のリスト)------->{{#def:MASS|{12, 1, 35, 19, 127, 14, 16, 31, 32} }} | <!---各原子の質量(上のリスト)------->{{#def:MASS|{12, 1, 35, 19, 127, 14, 16, 31, 32} }} | ||
| + | <!---化合物の名前------------------->{{#def:NAME|{{#nth:{{#var:dataline}}|6|&&}}}} | ||
| − | = | + | <span style="float:right;"> |
| − | {{ | + | [[Image:{{#var:MASSBANKID}}.png|150px]]<br/> |
| − | + | <big>{{#var:NAME}}</big><br/> | |
| + | </span> | ||
| + | |||
| + | <div> | ||
| + | {| class="wikitable nowraplinks" border="1" cellpadding="2" cellspacing="1" style="float: right; clear: none; margin: 1em 1em 1em 1em" | ||
| + | |- | ||
| + | ! colspan="2" | IDs and Links | ||
| + | |- class | ||
| + | | style="width: 35%;" | [http://massbank.jp/ MassBank] | ||
| + | | style="width: 65%;" | [http://www.massbank.jp/jsp/FwdRecord.jsp?id={{#var:MASSBANKID}} {{#var:NAME}}] | ||
| + | |- class{{#nth:{{#var:dataline}}|3|&&}}="hiddenStructure" | ||
| + | | style="width: 35%;" | [https://scifinder.cas.org/ CAS] | ||
| + | | style="width: 65%;" | {{#nth:{{#var:dataline}}|3|&&}} | ||
| + | |- class | ||
| + | | style="width: 35%;" | Keio ID | ||
| + | | style="width: 65%;" | {{#nth:{{#var:dataline}}|4|&&}} | ||
| + | |} | ||
| + | |||
| + | ==Top 10 Similar Molecules of {{#var:NAME}}== | ||
| + | {| class="wikitable collapsible collapsed" | ||
| + | ! Ranking | ||
| + | ! About scoring | ||
| + | |- | ||
| | | | ||
| − | + | {{#repeat:MassBank/Matrix/RankingLink|1| | |
| − | + | ||
{{#lua: | {{#lua: | ||
function intersection(x, y) | function intersection(x, y) | ||
| Line 35: | Line 58: | ||
total = 0 | total = 0 | ||
---Register Ions--- | ---Register Ions--- | ||
| + | ---全ページのイオンヘッダーにおける頻度を登録。最初の要素はページ名--- | ||
for id, frag in stdin:gmatch("&&([%a%d]+)&&([%S]+)") do | for id, frag in stdin:gmatch("&&([%a%d]+)&&([%S]+)") do | ||
table.insert(Name, id) | table.insert(Name, id) | ||
| Line 63: | Line 87: | ||
---Output Similarity--- | ---Output Similarity--- | ||
for i=table.getn(Score), math.max(table.getn(Score)-9,1), -1 do | for i=table.getn(Score), math.max(table.getn(Score)-9,1), -1 do | ||
| − | print( | + | print(Name[ Rank[Score[i]] ] .. " Score:" ..math.floor(Score[i])/100) |
end | end | ||
| | | | ||
| − | &&{{PAGENAME}}{{#car:{{{data}}}}} | + | &&{{PAGENAME}}&&{{#car:{{{data}}}}} |
{{#SearchLine:^&&_%&&|MassBank}} | {{#SearchLine:^&&_%&&|MassBank}} | ||
}} | }} | ||
| + | }} | ||
| + | |valign="top"| | ||
| + | The similarity score between two molecules is defined as the sum of Shannon entropy of ionic formulas shared by the molecules: | ||
| + | <math>\textstyle - \sum_{p\in shared} \log(\mbox{frequency}(p))</math> | ||
| + | <br/> | ||
| + | 代謝物間の類似度スコアは、それらのスペクトルで共有される(アノテートされた)イオン式のシャノン情報量の総和です。 | ||
| + | |} | ||
| + | |||
=Links= | =Links= | ||
| + | {| class="wikitable collapsible collapsed" | ||
| + | ! Page list for each fragment | ||
| + | |- | ||
| + | | | ||
{{#repeat:MassBank/SearchFgmt|1| | {{#repeat:MassBank/SearchFgmt|1| | ||
{{#lua: | {{#lua: | ||
| − | ---find and print Ruler--- | + | ---find and print Ruler--- |
| + | ---ヘッダー部分を分解するだけ--- | ||
for line in stdin:gmatch("[%S ,]+") do | for line in stdin:gmatch("[%S ,]+") do | ||
if (string.find(line, "^&&[&%a%d]+&&$") ~= nil) then | if (string.find(line, "^&&[&%a%d]+&&$") ~= nil) then | ||
| Line 81: | Line 118: | ||
|{{{data|CHClFINOPS}}} | |{{{data|CHClFINOPS}}} | ||
}}|&&}} | }}|&&}} | ||
| + | |} | ||
| − | =Annotations= | + | ==Annotations== |
| + | {{#ifeq:{{NAMESPACE}}|Ojima| | ||
| + | <div style="float:right"> | ||
| + | {{#ifexists:Image:Fragmentation:{{PAGENAME}}.png|[[Image:Fragmentation:{{PAGENAME}}.png]]}} | ||
| + | </div> | ||
| + | }} | ||
{{#replace: | {{#replace: | ||
{{#lua: | {{#lua: | ||
| Line 123: | Line 166: | ||
if (h ~= nil and t ~= nil) then | if (h ~= nil and t ~= nil) then | ||
if (comment ~= "") then | if (comment ~= "") then | ||
| − | ret = ret .."~-\n~~"..head.." ('''"..mass( | + | ret = ret .."~-\n~~"..head.." ('''"..mass(head).."''')".." ~~ "..tail.." ('''"..mass(tail).."''')".."\n~~ "..comment.."\n" |
comment = "" | comment = "" | ||
end | end | ||
| Line 145: | Line 188: | ||
|~|{{#bar:}}}} | |~|{{#bar:}}}} | ||
| − | =Precursor-Product Relationship= | + | ==Precursor-Product Relationship== |
| − | { | + | {| class="collapsible collapsed" |
| − | The matrix is viewed columnwise. The topmost precursor ion in bold face produces the product ions beneath it. | + | ! colspan="2"| About the PP Table (行列表示について) |
| + | |- | ||
| + | |The matrix is viewed columnwise. The topmost precursor ion in bold face produces the product ions beneath it. | ||
Each formula in matrix cells corresponds to the neutral loss. Blackout cells indicate products that cannot be derived, and orange cells indicate a structurally plausible link produced by cleaving a single chemical bond (in cases of ring-opening, two bonds). | Each formula in matrix cells corresponds to the neutral loss. Blackout cells indicate products that cannot be derived, and orange cells indicate a structurally plausible link produced by cleaving a single chemical bond (in cases of ring-opening, two bonds). | ||
| − | | | + | |行列は列方向に見ます。最上段太字の前駆イオン(precursor ion)が直下の生成イオン群(product ions)になると解釈します。 |
| − | 行列は列方向に見ます。最上段太字の前駆イオン(precursor ion)が直下の生成イオン群(product ions)になると解釈します。 | + | |
行列要素に書かれている組成式はニュートラルロスです。黒は前駆イオンから生成しえない関係、オレンジは分子構造における共有結合1本の切断(開環の場合は2本)で生じる関係を意味します。 | 行列要素に書かれている組成式はニュートラルロスです。黒は前駆イオンから生成しえない関係、オレンジは分子構造における共有結合1本の切断(開環の場合は2本)で生じる関係を意味します。 | ||
| − | + | |} | |
{{#replace: | {{#replace: | ||
{{#lua: | {{#lua: | ||
| Line 198: | Line 242: | ||
---Main Program--- | ---Main Program--- | ||
| − | local ruler = nil; | + | local ruler = nil; ---first line (list of fragments)--- |
| − | local fragments = {}; | + | local fragments = {}; ---following lines (head : tail style)--- |
| + | local focus = {}; ---formulas that should be highlighted--- | ||
local x, y; | local x, y; | ||
---Read Data--- | ---Read Data--- | ||
| Line 208: | Line 253: | ||
---register fragments--- | ---register fragments--- | ||
head, tail = string.match(line, "^&&(["..FORMULA_CHAR.."]+) *: *(["..FORMULA_CHAR.." ]+)$") | head, tail = string.match(line, "^&&(["..FORMULA_CHAR.."]+) *: *(["..FORMULA_CHAR.." ]+)$") | ||
| + | if (string.match(head,"*")) then | ||
| + | head = string.gsub(head,"*", "") | ||
| + | focus[head] = head | ||
| + | end | ||
if (head ~= nil and tail ~= nil) then | if (head ~= nil and tail ~= nil) then | ||
| − | if (fragments[head] == nil) then y = {} else y = fragments[head] end | + | if (fragments[head] == nil) |
| + | then y = {} else y = fragments[head] end | ||
for x in string.gmatch(tail,"["..FORMULA_CHAR.."]+") do | for x in string.gmatch(tail,"["..FORMULA_CHAR.."]+") do | ||
| − | y[x] = | + | z = string.gsub(x,"*", "") |
| + | y[x] = z | ||
| + | y[z] = z | ||
end | end | ||
fragments[head] = y | fragments[head] = y | ||
| Line 225: | Line 277: | ||
end | end | ||
x = nil | x = nil | ||
| − | + | inRuler = {} | |
for formula in string.gmatch(ruler, "([%S]+)") do | for formula in string.gmatch(ruler, "([%S]+)") do | ||
if (x ~= nil and mass(formula) > mass(x)) then | if (x ~= nil and mass(formula) > mass(x)) then | ||
| Line 231: | Line 283: | ||
end | end | ||
x = formula | x = formula | ||
| − | + | inRuler[formula] = true | |
end | end | ||
---Check Fragments--- | ---Check Fragments--- | ||
for i,v in pairs(fragments) do | for i,v in pairs(fragments) do | ||
| − | if (not | + | ---i is head, v is a list of fragments--- |
| + | if (not inRuler[i]) then print('<span style="color:red">Parent ion '..i..' does not exist.</span>') end | ||
for j,w in pairs(v) do | for j,w in pairs(v) do | ||
| − | if (not | + | j = string.gsub(j,"*","") |
| + | if (not inRuler[w]) then print('<span style="color:red">Fragment ion '..w..' does not exist.</span>') end | ||
end | end | ||
end | end | ||
---Write Matrix--- | ---Write Matrix--- | ||
print("<small>") | print("<small>") | ||
| − | print('{# class ="wikitable"') | + | print('{# class ="wikitable collapsible" style="margin:0 0 0 0"') |
local axis = {}; | local axis = {}; | ||
local i = 1; | local i = 1; | ||
| Line 251: | Line 305: | ||
---Header--- | ---Header--- | ||
print("#-\n") | print("#-\n") | ||
| − | print(" | + | print("! {{PAGENAME}} \n") |
for i=1, table.getn(axis) do | for i=1, table.getn(axis) do | ||
| − | print("#style='text-align:right'# '''" .. mass(axis[i]) .. "'''<br/>" .. axis[i]) | + | ---If some fragments are focused then highlight the axis--- |
| + | if (focus[axis[i]] ~= nil) then | ||
| + | print("#style='text-align:right;background:red'# '''" .. mass(axis[i]) .. "'''<br/>" .. axis[i]) | ||
| + | else | ||
| + | print("#style='text-align:right'# '''" .. mass(axis[i]) .. "'''<br/>" .. axis[i]) | ||
| + | end | ||
end | end | ||
---Rows--- | ---Rows--- | ||
for i=1, table.getn(axis) do | for i=1, table.getn(axis) do | ||
print("#-\n") | print("#-\n") | ||
| − | print("#style='text-align:right'# '''" .. mass(axis[i]) .. "''' | + | print("#style='text-align:right'# '''" .. mass(axis[i]) .. "''' " .. axis[i]) |
for j=1, table.getn(axis) do | for j=1, table.getn(axis) do | ||
if (j < i) then | if (j < i) then | ||
| Line 266: | Line 325: | ||
else | else | ||
if (fragments[axis[j]] ~= nil and fragments[axis[j]][axis[i]] ~= nil) then | if (fragments[axis[j]] ~= nil and fragments[axis[j]][axis[i]] ~= nil) then | ||
| − | print('#style="background-color:orange"# ' .. s) | + | ---記号の有無により色を変更--- |
| + | if (fragments[axis[j]][axis[i].."*"] ~= nil) then | ||
| + | print('#style="background-color:skyblue"# ' .. s) | ||
| + | elseif (fragments[axis[j]][axis[i].."**"] ~= nil) then | ||
| + | print('#style="background-color:orange"# ' .. s) | ||
| + | elseif (fragments[axis[j]][axis[i].."***"] ~= nil) then | ||
| + | print('#style="background-color:beige"# ' .. s) | ||
| + | elseif (fragments[axis[j]][axis[i].."****"] ~= nil) then | ||
| + | print('#style="background-color:coral"# ' .. s) | ||
| + | else | ||
| + | print('#style="background-color:orange"# ' .. s) | ||
| + | end | ||
else print('## ' .. s) end | else print('## ' .. s) end | ||
end | end | ||
| Line 283: | Line 353: | ||
<!---- next/prev links at the page bottom----> | <!---- next/prev links at the page bottom----> | ||
| + | {{#def:KeioID|{{#ifeq:{{#substring:{{PAGENAME}}|0|2}}|KO|{{#substring:{{#nth:{{#SearchLine:{{PAGENAME}}|Index|MassBank/Molecules}}|4|&&}}|0|4}}|{{#substring:{{PAGENAME}}|0|8}}}}}} | ||
{{#repeat:MassBank/PrevNextLink|4| | {{#repeat:MassBank/PrevNextLink|4| | ||
{{#lua: | {{#lua: | ||
| Line 290: | Line 361: | ||
last = nil | last = nil | ||
hit = nil | hit = nil | ||
| − | for | + | for word in stdin:gmatch("(%w+)") do |
| − | if (first == nil) then first = | + | if (first == nil) then first = word end |
| − | last = | + | last = word |
| − | if ( | + | if (word == "{{#var:KeioID}}") then |
hit = true | hit = true | ||
if (tmp ~= nil) then | if (tmp ~= nil) then | ||
| Line 301: | Line 372: | ||
end | end | ||
elseif (hit) then | elseif (hit) then | ||
| − | next = | + | next = word |
hit = false | hit = false | ||
end | end | ||
| − | tmp = | + | tmp = word |
end | end | ||
if (next == nil) then next = last end | if (next == nil) then next = last end | ||
Latest revision as of 16:27, 7 April 2011
| General Index | Ion Frequency | Prec.-Product | Neutral Loss | Help |
| IDs and Links | |
|---|---|
| MassBank | [1] |
| CAS | |
| Keio ID | |
Contents |
[edit] Top 10 Similar Molecules of
| Ranking | About scoring |
|---|---|
|
The similarity score between two molecules is defined as the sum of Shannon entropy of ionic formulas shared by the molecules:
|
[edit] Links
| Page list for each fragment |
|---|
[edit] Annotations
| Precursor | Product | Comments |
|---|---|---|
| (0) | (0) | CHClFINOPS |
[edit] Precursor-Product Relationship
| About the PP Table (行列表示について) | |
|---|---|
| The matrix is viewed columnwise. The topmost precursor ion in bold face produces the product ions beneath it.
Each formula in matrix cells corresponds to the neutral loss. Blackout cells indicate products that cannot be derived, and orange cells indicate a structurally plausible link produced by cleaving a single chemical bond (in cases of ring-opening, two bonds). |
行列は列方向に見ます。最上段太字の前駆イオン(precursor ion)が直下の生成イオン群(product ions)になると解釈します。
行列要素に書かれている組成式はニュートラルロスです。黒は前駆イオンから生成しえない関係、オレンジは分子構造における共有結合1本の切断(開環の場合は2本)で生じる関係を意味します。 |
No "ION INFO" line in the form &&(formula)&&(formula)&&...&&
| MassBank/Matrix | 210 CHClFIONSP |
|---|---|
| 210 CHClFIONSP |
|