Mol:FLNAACGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.8320 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8320 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5508 2.5121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5508 2.5121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2693 2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2693 2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2693 1.2369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2693 1.2369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0146 0.8066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0146 0.8066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7597 1.2369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7597 1.2369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7597 2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7597 2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0146 2.5275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0146 2.5275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8320 1.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8320 1.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5508 0.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5508 0.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0146 -0.0533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0146 -0.0533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3000 -0.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3000 -0.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3000 -1.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3000 -1.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0146 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0146 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7290 -1.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7290 -1.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7290 -0.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7290 -0.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5045 2.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5045 2.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1134 2.5120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1134 2.5120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0146 -2.5275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0146 -2.5275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5537 0.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5537 0.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9305 -0.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9305 -0.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4677 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4677 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1988 -0.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1988 -0.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8418 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8418 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3746 0.0948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3746 0.0948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3062 -0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3062 -0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6119 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6119 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1260 -0.6340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1260 -0.6340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7883 -0.7950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7883 -0.7950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0160 0.4024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0160 0.4024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7392 0.4040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7392 0.4040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4428 -1.7029 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4428 -1.7029 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6980 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6980 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4368 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4368 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1067 1.2422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1067 1.2422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0631 0.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0631 0.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9971 1.1089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9971 1.1089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4368 2.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4368 2.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6966 2.1916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6966 2.1916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4385 1.1958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4385 1.1958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5045 1.1958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5045 1.1958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 1 9 1 0 0 0 0 | + | 1 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 1 0 0 0 0 | + | 5 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 10 20 1 0 0 0 0 | + | 10 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 15 32 1 0 0 0 0 | + | 15 32 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 33 1 0 0 0 0 | + | 37 33 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 37 31 1 0 0 0 0 | + | 37 31 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 0.0017 0.4899 | + | M SBV 1 45 0.0017 0.4899 |
| − | S SKP 5 | + | S SKP 5 |
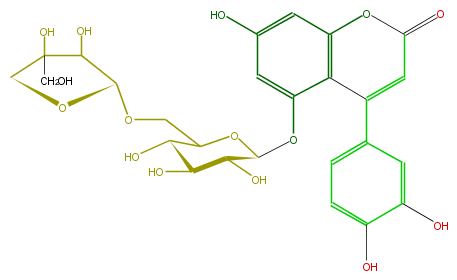
| − | ID FLNAACGS0002 | + | ID FLNAACGS0002 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES c(c3)(O5)c(C(=CC5=O)c(c4)ccc(c(O)4)O)c(cc3O)OC(O1)C(O)C(C(C(COC(C2O)OCC2(CO)O)1)O)O | + | SMILES c(c3)(O5)c(C(=CC5=O)c(c4)ccc(c(O)4)O)c(cc3O)OC(O1)C(O)C(C(C(COC(C2O)OCC2(CO)O)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
0.8320 2.0971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5508 2.5121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2693 2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2693 1.2369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0146 0.8066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7597 1.2369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7597 2.0974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0146 2.5275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8320 1.2676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5508 0.8528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0146 -0.0533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3000 -0.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3000 -1.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0146 -1.7032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7290 -1.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7290 -0.4657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5045 2.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1134 2.5120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0146 -2.5275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5537 0.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9305 -0.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4677 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1988 -0.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8418 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3746 0.0948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3062 -0.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6119 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1260 -0.6340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7883 -0.7950 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0160 0.4024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7392 0.4040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4428 -1.7029 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6980 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4368 1.6857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1067 1.2422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0631 0.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9971 1.1089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4368 2.1678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6966 2.1916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4385 1.1958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5045 1.1958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
1 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
1 18 1 0 0 0 0
14 19 1 0 0 0 0
10 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 30 1 0 0 0 0
24 20 1 0 0 0 0
30 31 1 0 0 0 0
15 32 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 33 1 0 0 0 0
34 38 1 0 0 0 0
33 39 1 0 0 0 0
37 31 1 0 0 0 0
40 41 1 0 0 0 0
34 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 0.0017 0.4899
S SKP 5
ID FLNAACGS0002
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES c(c3)(O5)c(C(=CC5=O)c(c4)ccc(c(O)4)O)c(cc3O)OC(O1)C(O)C(C(C(COC(C2O)OCC2(CO)O)1)O)O
M END