Mol:FLID1AGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 41 0 0 0 0 0 0 0 0999 V2000 | + | 37 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0568 -0.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0568 -0.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5005 -0.4263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5005 -0.4263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0558 -0.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0558 -0.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6119 -0.4262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6119 -0.4262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6119 -1.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6119 -1.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1887 -1.4252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1887 -1.4252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7654 -1.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7654 -1.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7654 -0.4262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7654 -0.4262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1887 -0.0932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1887 -0.0932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5002 -1.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5002 -1.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0558 -1.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0558 -1.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1925 -2.0616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1925 -2.0616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3418 -1.4249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3418 -1.4249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3418 -2.0616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3418 -2.0616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8931 -2.3799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8931 -2.3799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4445 -2.0616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4445 -2.0616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4445 -1.4249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4445 -1.4249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8931 -1.1066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8931 -1.1066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1607 -0.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1607 -0.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8144 -0.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8144 -0.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3158 -0.5721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3158 -0.5721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7962 -0.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7962 -0.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1843 -0.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1843 -0.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6936 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6936 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8589 -0.4960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8589 -0.4960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3542 -0.7923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3542 -0.7923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0301 -1.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0301 -1.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1786 0.0849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1786 0.0849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5819 0.6197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5819 0.6197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6011 1.4655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6011 1.4655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1626 1.7897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1626 1.7897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8699 1.8877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8699 1.8877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8699 2.5791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8699 2.5791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3384 2.8496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3384 2.8496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2722 2.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2722 2.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8589 -2.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8589 -2.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0620 -2.7107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0620 -2.7107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 1 0 0 0 0 | + | 6 12 1 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 14 12 1 0 0 0 0 | + | 14 12 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 30 29 1 0 0 0 0 | + | 30 29 1 0 0 0 0 |
| − | 16 36 1 0 0 0 0 | + | 16 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 36 37 | + | M SAL 1 2 36 37 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 40 -5.7108 3.4283 | + | M SBV 1 40 -5.7108 3.4283 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLID1AGS0002 | + | ID FLID1AGS0002 |
| − | KNApSAcK_ID C00010184 | + | KNApSAcK_ID C00010184 |
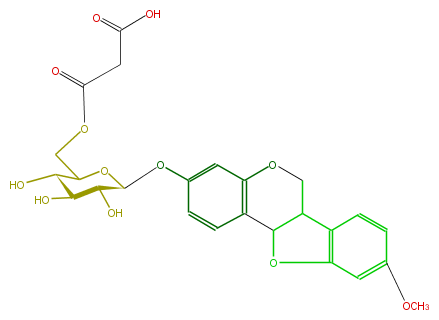
| − | NAME Medicarpin 3-O-(6'-malonylgluclside) | + | NAME Medicarpin 3-O-(6'-malonylgluclside) |
| − | CAS_RN 131653-24-2 | + | CAS_RN 131653-24-2 |
| − | FORMULA C25H26O12 | + | FORMULA C25H26O12 |
| − | EXACTMASS 518.1424262959999 | + | EXACTMASS 518.1424262959999 |
| − | AVERAGEMASS 518.46674 | + | AVERAGEMASS 518.46674 |
| − | SMILES O=C(CC(O)=O)OCC(C(O)1)OC(Oc(c5)cc(c(c5)2)OCC(c34)C(Oc3cc(cc4)OC)2)C(C1O)O | + | SMILES O=C(CC(O)=O)OCC(C(O)1)OC(Oc(c5)cc(c(c5)2)OCC(c34)C(Oc3cc(cc4)OC)2)C(C1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 41 0 0 0 0 0 0 0 0999 V2000
-1.0568 -0.1052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5005 -0.4263 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0558 -0.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6119 -0.4262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6119 -1.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1887 -1.4252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7654 -1.0922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7654 -0.4262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1887 -0.0932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5002 -1.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0558 -1.3894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1925 -2.0616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3418 -1.4249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3418 -2.0616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8931 -2.3799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4445 -2.0616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4445 -1.4249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8931 -1.1066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1607 -0.3089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8144 -0.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3158 -0.5721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7962 -0.5778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1843 -0.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6936 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8589 -0.4960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3542 -0.7923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0301 -1.0517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1786 0.0849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5819 0.6197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6011 1.4655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1626 1.7897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8699 1.8877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8699 2.5791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3384 2.8496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2722 2.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8589 -2.9243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0620 -2.7107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 1 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 1 1 0 0 0 0
14 12 1 0 0 0 0
24 28 1 0 0 0 0
28 29 1 0 0 0 0
30 31 2 0 0 0 0
30 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
30 29 1 0 0 0 0
16 36 1 0 0 0 0
36 37 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 36 37
M SBL 1 1 40
M SMT 1 OCH3
M SBV 1 40 -5.7108 3.4283
S SKP 8
ID FLID1AGS0002
KNApSAcK_ID C00010184
NAME Medicarpin 3-O-(6'-malonylgluclside)
CAS_RN 131653-24-2
FORMULA C25H26O12
EXACTMASS 518.1424262959999
AVERAGEMASS 518.46674
SMILES O=C(CC(O)=O)OCC(C(O)1)OC(Oc(c5)cc(c(c5)2)OCC(c34)C(Oc3cc(cc4)OC)2)C(C1O)O
M END