Mol:FLIACFGM0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0254 1.7100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0254 1.7100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3046 2.1260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3046 2.1260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5840 1.7101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5840 1.7101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5840 0.8471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5840 0.8471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1632 0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1632 0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9106 0.8471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9106 0.8471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9106 1.7101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9106 1.7101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1632 2.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1632 2.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0251 0.8781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0251 0.8781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3046 0.4620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3046 0.4620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6574 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6574 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6574 -0.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6574 -0.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3718 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3718 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0862 -0.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0862 -0.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0862 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0862 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3718 0.8285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3718 0.8285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3046 -0.4093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3046 -0.4093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1632 -0.4467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1632 -0.4467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6733 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6733 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2275 -1.0884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2275 -1.0884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5854 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5854 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9658 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9658 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4161 -0.3817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4161 -0.3817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0719 -0.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0719 -0.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3559 -0.7497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3559 -0.7497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9151 -1.0456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9151 -1.0456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9273 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9273 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3046 2.8508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3046 2.8508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1180 -2.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1180 -2.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4655 -2.7106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4655 -2.7106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7972 -2.5196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7972 -2.5196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1248 -2.7106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1248 -2.7106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7772 -2.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7772 -2.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4457 -2.2996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4457 -2.2996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8329 -2.0780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8329 -2.0780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9834 -2.4871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9834 -2.4871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0577 -3.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0577 -3.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1248 -3.2972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1248 -3.2972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8310 2.1751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8310 2.1751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4789 3.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4789 3.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7355 -0.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7355 -0.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9894 0.2610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9894 0.2610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8007 0.8285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8007 0.8285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7263 0.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7263 0.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8673 -0.8599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8673 -0.8599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9894 -1.5079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9894 -1.5079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 2 0 0 0 0 | + | 6 7 2 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 1 9 1 0 0 0 0 | + | 1 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 6 11 1 0 0 0 0 | + | 6 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 10 17 1 0 0 0 0 | + | 10 17 1 0 0 0 0 |
| − | 5 18 2 0 0 0 0 | + | 5 18 2 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 17 1 0 0 0 0 | + | 22 17 1 0 0 0 0 |
| − | 2 28 1 0 0 0 0 | + | 2 28 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 27 1 0 0 0 0 | + | 33 27 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 1 39 1 0 0 0 0 | + | 1 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 14 45 1 0 0 0 0 | + | 14 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 44 0.8056 -0.4651 | + | M SBV 1 44 0.8056 -0.4651 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 46 0.6637 -0.5517 | + | M SBV 2 46 0.6637 -0.5517 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 48 -0.7145 -0.4124 | + | M SBV 3 48 -0.7145 -0.4124 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 45 46 | + | M SAL 4 2 45 46 |
| − | M SBL 4 1 50 | + | M SBL 4 1 50 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SBV 4 50 -0.7811 0.4510 | + | M SBV 4 50 -0.7811 0.4510 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FLIACFGM0001 | + | ID FLIACFGM0001 |
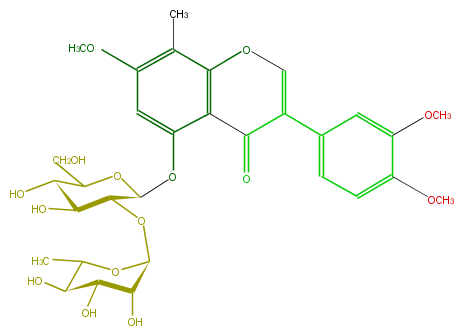
| − | FORMULA C31H38O15 | + | FORMULA C31H38O15 |
| − | EXACTMASS 650.221070546 | + | EXACTMASS 650.221070546 |
| − | AVERAGEMASS 650.62442 | + | AVERAGEMASS 650.62442 |
| − | SMILES O(C(C5O)C(OC(C(O)5)CO)Oc(c32)cc(c(C)c(OC=C(c(c4)cc(OC)c(OC)c4)C3=O)2)OC)C(O1)C(O)C(C(O)C1C)O | + | SMILES O(C(C5O)C(OC(C(O)5)CO)Oc(c32)cc(c(C)c(OC=C(c(c4)cc(OC)c(OC)c4)C3=O)2)OC)C(O1)C(O)C(C(O)C1C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-2.0254 1.7100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3046 2.1260 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5840 1.7101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5840 0.8471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1632 0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9106 0.8471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9106 1.7101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1632 2.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0251 0.8781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3046 0.4620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6574 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6574 -0.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3718 -0.8214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0862 -0.4090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0862 0.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3718 0.8285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3046 -0.4093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1632 -0.4467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6733 -0.4999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2275 -1.0884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5854 -0.8387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9658 -0.8320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4161 -0.3817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0719 -0.6173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3559 -0.7497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9151 -1.0456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9273 -1.2926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3046 2.8508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1180 -2.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4655 -2.7106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7972 -2.5196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1248 -2.7106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7772 -2.1087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4457 -2.2996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8329 -2.0780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9834 -2.4871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0577 -3.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1248 -3.2972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8310 2.1751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4789 3.2972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7355 -0.0655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9894 0.2610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8007 0.8285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7263 0.2940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8673 -0.8599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9894 -1.5079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
1 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
6 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
10 17 1 0 0 0 0
5 18 2 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 17 1 0 0 0 0
2 28 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 27 1 0 0 0 0
39 40 1 0 0 0 0
1 39 1 0 0 0 0
41 42 1 0 0 0 0
24 41 1 0 0 0 0
43 44 1 0 0 0 0
15 43 1 0 0 0 0
45 46 1 0 0 0 0
14 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 ^ OCH3
M SBV 1 44 0.8056 -0.4651
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 ^ CH2OH
M SBV 2 46 0.6637 -0.5517
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 48
M SMT 3 OCH3
M SBV 3 48 -0.7145 -0.4124
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 45 46
M SBL 4 1 50
M SMT 4 OCH3
M SBV 4 50 -0.7811 0.4510
S SKP 5
ID FLIACFGM0001
FORMULA C31H38O15
EXACTMASS 650.221070546
AVERAGEMASS 650.62442
SMILES O(C(C5O)C(OC(C(O)5)CO)Oc(c32)cc(c(C)c(OC=C(c(c4)cc(OC)c(OC)c4)C3=O)2)OC)C(O1)C(O)C(C(O)C1C)O
M END