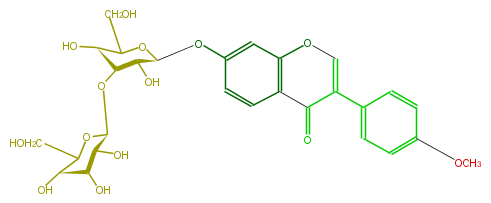
Mol:FLIA1AGS0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4504 0.9761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4504 0.9761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1059 0.6549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1059 0.6549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6622 0.9761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6622 0.9761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2183 0.6551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2183 0.6551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2183 -0.0109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2183 -0.0109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7951 -0.3439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7951 -0.3439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3718 -0.0109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3718 -0.0109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3718 0.6551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3718 0.6551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7951 0.9880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7951 0.9880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1061 0.0129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1061 0.0129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6622 -0.3081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6622 -0.3081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7951 -1.0094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7951 -1.0094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9481 -0.3436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9481 -0.3436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9481 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9481 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4995 -1.2986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4995 -1.2986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0508 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0508 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0508 -0.3436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0508 -0.3436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4995 -0.0253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4995 -0.0253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5556 0.8864 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.5556 0.8864 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.1497 0.4391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1497 0.4391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.7138 0.6087 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.7138 0.6087 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2932 0.6132 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2932 0.6132 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5988 0.9189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5988 0.9189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0441 0.7590 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.0441 0.7590 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.0193 0.9270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0193 0.9270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4333 0.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4333 0.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4640 0.1894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4640 0.1894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3440 -1.7001 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.3440 -1.7001 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.7405 -1.6766 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7405 -1.6766 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5902 -1.2337 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5902 -1.2337 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3233 -0.9086 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3233 -0.9086 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.7539 -0.9462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7539 -0.9462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9177 -1.3901 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9177 -1.3901 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.6731 -2.0293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6731 -2.0293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4874 -2.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4874 -2.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1084 -1.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1084 -1.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9168 -1.4803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9168 -1.4803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7828 -1.9804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7828 -1.9804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3164 1.5990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3164 1.5990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6468 2.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6468 2.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7362 -1.0589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7362 -1.0589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5246 -1.6740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5246 -1.6740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 16 37 1 0 0 0 0 | + | 16 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 24 39 1 0 0 0 0 | + | 24 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 41 42 | + | M SAL 3 2 41 42 |
| − | M SBL 3 1 45 | + | M SBL 3 1 45 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 45 -4.317 0.6915 | + | M SVB 3 45 -4.317 0.6915 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 39 40 | + | M SAL 2 2 39 40 |
| − | M SBL 2 1 43 | + | M SBL 2 1 43 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 43 -2.3164 1.599 | + | M SVB 2 43 -2.3164 1.599 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 41 4.2449 -1.1865 | + | M SVB 1 41 4.2449 -1.1865 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIA1AGS0007 | + | ID FLIA1AGS0007 |
| − | KNApSAcK_ID C00010082 | + | KNApSAcK_ID C00010082 |
| − | NAME Formononetin 7-O-laminaribioside | + | NAME Formononetin 7-O-laminaribioside |
| − | CAS_RN 56222-47-0 | + | CAS_RN 56222-47-0 |
| − | FORMULA C28H32O14 | + | FORMULA C28H32O14 |
| − | EXACTMASS 592.179205732 | + | EXACTMASS 592.179205732 |
| − | AVERAGEMASS 592.54528 | + | AVERAGEMASS 592.54528 |
| − | SMILES [C@H](O[C@H]([C@@H]2O)[C@@H](O)C(CO)O[C@H]2Oc(c5)ccc(c35)C(C(c(c4)ccc(c4)OC)=CO3)=O)(O1)[C@H]([C@H]([C@H](C1CO)O)O)O | + | SMILES [C@H](O[C@H]([C@@H]2O)[C@@H](O)C(CO)O[C@H]2Oc(c5)ccc(c35)C(C(c(c4)ccc(c4)OC)=CO3)=O)(O1)[C@H]([C@H]([C@H](C1CO)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.4504 0.9761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1059 0.6549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6622 0.9761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2183 0.6551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2183 -0.0109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7951 -0.3439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3718 -0.0109 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3718 0.6551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7951 0.9880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1061 0.0129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6622 -0.3081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7951 -1.0094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9481 -0.3436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9481 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4995 -1.2986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0508 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0508 -0.3436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4995 -0.0253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5556 0.8864 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.1497 0.4391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7138 0.6087 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2932 0.6132 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5988 0.9189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0441 0.7590 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.0193 0.9270 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4333 0.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4640 0.1894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3440 -1.7001 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.7405 -1.6766 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5902 -1.2337 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3233 -0.9086 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7539 -0.9462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9177 -1.3901 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.6731 -2.0293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4874 -2.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1084 -1.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9168 -1.4803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7828 -1.9804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3164 1.5990 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6468 2.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7362 -1.0589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5246 -1.6740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 1 1 0 0 0 0
28 29 1 1 0 0 0
29 30 1 1 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 26 1 0 0 0 0
16 37 1 0 0 0 0
37 38 1 0 0 0 0
24 39 1 0 0 0 0
39 40 1 0 0 0 0
33 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 41 42
M SBL 3 1 45
M SMT 3 CH2OH
M SVB 3 45 -4.317 0.6915
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 39 40
M SBL 2 1 43
M SMT 2 CH2OH
M SVB 2 43 -2.3164 1.599
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 41
M SMT 1 OCH3
M SVB 1 41 4.2449 -1.1865
S SKP 8
ID FLIA1AGS0007
KNApSAcK_ID C00010082
NAME Formononetin 7-O-laminaribioside
CAS_RN 56222-47-0
FORMULA C28H32O14
EXACTMASS 592.179205732
AVERAGEMASS 592.54528
SMILES [C@H](O[C@H]([C@@H]2O)[C@@H](O)C(CO)O[C@H]2Oc(c5)ccc(c35)C(C(c(c4)ccc(c4)OC)=CO3)=O)(O1)[C@H]([C@H]([C@H](C1CO)O)O)O
M END