Mol:FLIA1AGI0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 51 0 0 0 0 0 0 0 0999 V2000 | + | 47 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7282 1.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7282 1.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1719 0.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1719 0.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3844 1.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3844 1.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9405 0.9346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9405 0.9346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9405 0.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9405 0.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5173 -0.0644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5173 -0.0644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0940 0.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0940 0.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0940 0.9346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0940 0.9346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5173 1.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5173 1.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1717 0.2924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1717 0.2924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3844 -0.0286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3844 -0.0286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5173 -0.7299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5173 -0.7299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6704 -0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6704 -0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6704 -0.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6704 -0.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2217 -1.0191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2217 -1.0191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7731 -0.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7731 -0.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7731 -0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7731 -0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2217 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2217 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8321 1.0519 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.8321 1.0519 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.4858 0.5948 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4858 0.5948 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9872 0.7887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9872 0.7887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.4676 0.7830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.4676 0.7830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8557 1.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8557 1.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3650 0.9607 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.3650 0.9607 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.5303 0.8648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5303 0.8648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0256 0.5685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0256 0.5685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7016 0.3091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7016 0.3091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7016 -0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7016 -0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2083 -0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2083 -0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3803 -0.8678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3803 -0.8678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0579 -0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0579 -0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7342 -0.8671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7342 -0.8671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2984 -0.5414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2984 -0.5414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8626 -0.8671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8626 -0.8671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8626 -1.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8626 -1.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2984 -1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2984 -1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7342 -1.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7342 -1.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4259 -1.8438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4259 -1.8438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3244 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3244 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3244 0.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3244 0.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8757 1.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8757 1.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4259 0.8915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4259 0.8915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8757 1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8757 1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6391 -1.2008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6391 -1.2008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5051 -1.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5051 -1.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3032 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3032 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4725 2.2924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4725 2.2924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 2 0 0 0 0 | + | 28 29 2 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 17 39 1 0 0 0 0 | + | 17 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 24 46 1 0 0 0 0 | + | 24 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 46 47 | + | M SAL 2 2 46 47 |
| − | M SBL 2 1 50 | + | M SBL 2 1 50 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 50 -2.1073 1.2998 | + | M SVB 2 50 -2.1073 1.2998 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 44 45 | + | M SAL 1 2 44 45 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 48 3.9672 -0.907 | + | M SVB 1 48 3.9672 -0.907 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIA1AGI0001 | + | ID FLIA1AGI0001 |
| − | KNApSAcK_ID C00010173 | + | KNApSAcK_ID C00010173 |
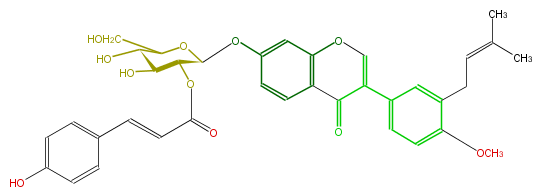
| − | NAME 4'-O-Methylneobavaisoflavone 7-O-(2''-p-coumaroylglucoside) | + | NAME 4'-O-Methylneobavaisoflavone 7-O-(2''-p-coumaroylglucoside) |
| − | CAS_RN 126654-66-8 | + | CAS_RN 126654-66-8 |
| − | FORMULA C36H36O11 | + | FORMULA C36H36O11 |
| − | EXACTMASS 644.225761994 | + | EXACTMASS 644.225761994 |
| − | AVERAGEMASS 644.66444 | + | AVERAGEMASS 644.66444 |
| − | SMILES O(C(C=Cc(c5)ccc(O)c5)=O)[C@@H]([C@@H](O)1)[C@H](Oc(c2)ccc(C(=O)3)c2OC=C3c(c4)cc(c(OC)c4)CC=C(C)C)OC(CO)[C@@H]1O | + | SMILES O(C(C=Cc(c5)ccc(O)c5)=O)[C@@H]([C@@H](O)1)[C@H](Oc(c2)ccc(C(=O)3)c2OC=C3c(c4)cc(c(OC)c4)CC=C(C)C)OC(CO)[C@@H]1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 51 0 0 0 0 0 0 0 0999 V2000
-0.7282 1.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1719 0.9344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3844 1.2556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9405 0.9346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9405 0.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5173 -0.0644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0940 0.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0940 0.9346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5173 1.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1717 0.2924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3844 -0.0286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5173 -0.7299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6704 -0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6704 -0.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2217 -1.0191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7731 -0.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7731 -0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2217 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8321 1.0519 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.4858 0.5948 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9872 0.7887 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.4676 0.7830 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8557 1.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3650 0.9607 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5303 0.8648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0256 0.5685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7016 0.3091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7016 -0.4759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2083 -0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3803 -0.8678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0579 -0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7342 -0.8671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2984 -0.5414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8626 -0.8671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8626 -1.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2984 -1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7342 -1.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4259 -1.8438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3244 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3244 0.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8757 1.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4259 0.8915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8757 1.8444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6391 -1.2008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5051 -1.7008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3032 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4725 2.2924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 1 1 0 0 0 0
27 28 1 0 0 0 0
28 29 2 0 0 0 0
28 30 1 0 0 0 0
30 31 2 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 32 1 0 0 0 0
35 38 1 0 0 0 0
17 39 1 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
41 43 1 0 0 0 0
16 44 1 0 0 0 0
44 45 1 0 0 0 0
24 46 1 0 0 0 0
46 47 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 46 47
M SBL 2 1 50
M SMT 2 CH2OH
M SVB 2 50 -2.1073 1.2998
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 44 45
M SBL 1 1 48
M SMT 1 OCH3
M SVB 1 48 3.9672 -0.907
S SKP 8
ID FLIA1AGI0001
KNApSAcK_ID C00010173
NAME 4'-O-Methylneobavaisoflavone 7-O-(2''-p-coumaroylglucoside)
CAS_RN 126654-66-8
FORMULA C36H36O11
EXACTMASS 644.225761994
AVERAGEMASS 644.66444
SMILES O(C(C=Cc(c5)ccc(O)c5)=O)[C@@H]([C@@H](O)1)[C@H](Oc(c2)ccc(C(=O)3)c2OC=C3c(c4)cc(c(OC)c4)CC=C(C)C)OC(CO)[C@@H]1O
M END