Mol:FL7ARXGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 44 0 0 0 0 0 0 0 0999 V2000 | + | 40 44 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3533 -2.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -2.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6496 -1.6414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6496 -1.6414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6496 -0.8288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6496 -0.8288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0569 -0.8288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0569 -0.8288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0569 -1.6414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0569 -1.6414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0569 0.7962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0569 0.7962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7606 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7606 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7606 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7606 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0541 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0541 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0541 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0541 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6496 0.7962 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.6496 0.7962 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8374 0.8422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8374 0.8422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 0.4438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 0.4438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2176 0.8422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2176 0.8422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2176 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2176 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 2.0375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 2.0375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8374 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8374 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3337 0.7209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3337 0.7209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3533 -2.6920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3533 -2.6920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9026 -3.0092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9026 -3.0092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7671 -3.0304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7671 -3.0304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7207 -0.8074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7207 -0.8074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5275 2.5724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5275 2.5724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9855 3.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9855 3.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6259 1.8748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6259 1.8748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6733 0.5791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6733 0.5791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3337 0.5791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3337 0.5791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0666 -0.5036 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.0666 -0.5036 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.2491 -0.6142 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2491 -0.6142 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.9136 -1.1031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9136 -1.1031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2362 -1.8625 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2362 -1.8625 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0538 -1.7519 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0538 -1.7519 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3893 -1.2629 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3893 -1.2629 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.3600 -2.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3600 -2.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5625 -3.4582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5625 -3.4582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8997 -2.2852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8997 -2.2852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9078 -1.1928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9078 -1.1928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5132 -0.1841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5132 -0.1841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 3 11 2 0 0 0 0 | + | 3 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 7 1 0 0 0 0 | + | 13 7 1 0 0 0 0 |
| − | 12 14 1 0 0 0 0 | + | 12 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 9 20 1 0 0 0 0 | + | 9 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 21 22 2 0 0 0 0 | + | 21 22 2 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 11 24 1 0 0 0 0 | + | 11 24 1 0 0 0 0 |
| − | 18 25 1 0 0 0 0 | + | 18 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 17 27 1 0 0 0 0 | + | 17 27 1 0 0 0 0 |
| − | 16 28 1 0 0 0 0 | + | 16 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 30 40 1 0 0 0 0 | + | 30 40 1 0 0 0 0 |
| − | 31 24 1 0 0 0 0 | + | 31 24 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 36 37 | + | M SAL 4 2 36 37 |
| − | M SBL 4 1 39 | + | M SBL 4 1 39 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 39 2.8497 -2.114 | + | M SVB 4 39 2.8497 -2.114 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 28 29 | + | M SAL 3 2 28 29 |
| − | M SBL 3 1 31 | + | M SBL 3 1 31 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 31 2.6733 0.5791 | + | M SVB 3 31 2.6733 0.5791 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 25 26 | + | M SAL 2 2 25 26 |
| − | M SBL 2 1 28 | + | M SBL 2 1 28 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 28 1.5275 2.5724 | + | M SVB 2 28 1.5275 2.5724 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 21 22 23 | + | M SAL 1 3 21 22 23 |
| − | M SBL 1 1 24 | + | M SBL 1 1 24 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 24 -1.3533 -2.692 | + | M SVB 1 24 -1.3533 -2.692 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7ARXGL0003 | + | ID FL7ARXGL0003 |
| − | KNApSAcK_ID C00011186 | + | KNApSAcK_ID C00011186 |
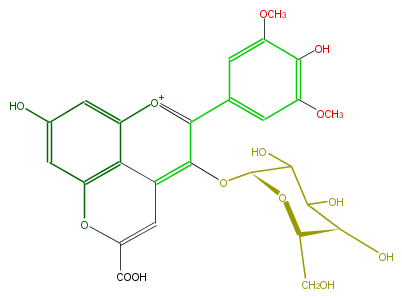
| − | NAME Malvidin 3-glucoside-pyruvate | + | NAME Malvidin 3-glucoside-pyruvate |
| − | CAS_RN 388089-38-1 | + | CAS_RN 388089-38-1 |
| − | FORMULA C26H25O14 | + | FORMULA C26H25O14 |
| − | EXACTMASS 561.124430508 | + | EXACTMASS 561.124430508 |
| − | AVERAGEMASS 561.4683 | + | AVERAGEMASS 561.4683 |
| − | SMILES c(O)(c(OC)1)c(OC)cc(c(c4O[C@@H](C5O)O[C@H](CO)[C@@H](C(O)5)O)[o+1]c(c3)c(c42)c(cc3O)OC(C(O)=O)=C2)c1 | + | SMILES c(O)(c(OC)1)c(OC)cc(c(c4O[C@@H](C5O)O[C@H](CO)[C@@H](C(O)5)O)[o+1]c(c3)c(c42)c(cc3O)OC(C(O)=O)=C2)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 44 0 0 0 0 0 0 0 0999 V2000
-1.3533 -2.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6496 -1.6414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6496 -0.8288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0569 -0.8288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0569 -1.6414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0569 0.7962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7606 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7606 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0541 -0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0541 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6496 0.7962 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.8374 0.8422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 0.4438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2176 0.8422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2176 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 2.0375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8374 1.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3337 0.7209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3533 -2.6920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9026 -3.0092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7671 -3.0304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7207 -0.8074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5275 2.5724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9855 3.0304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6259 1.8748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6733 0.5791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3337 0.5791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0666 -0.5036 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.2491 -0.6142 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9136 -1.1031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2362 -1.8625 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0538 -1.7519 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3893 -1.2629 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.3600 -2.8548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5625 -3.4582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8997 -2.2852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9078 -1.1928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5132 -0.1841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
3 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 7 1 0 0 0 0
12 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
9 20 1 0 0 0 0
1 21 1 0 0 0 0
21 22 2 0 0 0 0
21 23 1 0 0 0 0
11 24 1 0 0 0 0
18 25 1 0 0 0 0
25 26 1 0 0 0 0
17 27 1 0 0 0 0
16 28 1 0 0 0 0
28 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
33 36 1 0 0 0 0
36 37 1 0 0 0 0
34 38 1 0 0 0 0
35 39 1 0 0 0 0
30 40 1 0 0 0 0
31 24 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 36 37
M SBL 4 1 39
M SMT 4 CH2OH
M SVB 4 39 2.8497 -2.114
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 28 29
M SBL 3 1 31
M SMT 3 OCH3
M SVB 3 31 2.6733 0.5791
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 25 26
M SBL 2 1 28
M SMT 2 OCH3
M SVB 2 28 1.5275 2.5724
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 21 22 23
M SBL 1 1 24
M SMT 1 COOH
M SVB 1 24 -1.3533 -2.692
S SKP 8
ID FL7ARXGL0003
KNApSAcK_ID C00011186
NAME Malvidin 3-glucoside-pyruvate
CAS_RN 388089-38-1
FORMULA C26H25O14
EXACTMASS 561.124430508
AVERAGEMASS 561.4683
SMILES c(O)(c(OC)1)c(OC)cc(c(c4O[C@@H](C5O)O[C@H](CO)[C@@H](C(O)5)O)[o+1]c(c3)c(c42)c(cc3O)OC(C(O)=O)=C2)c1
M END