Mol:FL7AAIGL0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 57 62 0 0 0 0 0 0 0 0999 V2000 | + | 57 62 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.7223 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7223 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7223 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7223 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1660 0.4919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1660 0.4919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6097 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6097 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6097 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6097 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1660 1.7766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1660 1.7766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0534 0.4919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0534 0.4919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4971 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4971 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4971 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4971 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0534 1.7766 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -1.0534 1.7766 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0590 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0590 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6260 1.4491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6260 1.4491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1930 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1930 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1930 2.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1930 2.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6260 2.7585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6260 2.7585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0590 2.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0590 2.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2784 1.7765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2784 1.7765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0590 2.9312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0590 2.9312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1660 -0.1502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1660 -0.1502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0458 0.0314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0458 0.0314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4889 0.2274 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.4889 0.2274 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.1881 -0.2936 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.1881 -0.2936 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7666 -0.1283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7666 -0.1283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3486 -0.2936 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3486 -0.2936 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6494 0.2274 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6494 0.2274 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0709 0.0622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.0709 0.0622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9302 0.0777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9302 0.0777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1125 0.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1125 0.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0038 0.5818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0038 0.5818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9001 -0.2936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9001 -0.2936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2784 -0.9488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2784 -0.9488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0840 -2.7285 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 2.0840 -2.7285 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.6661 -2.5783 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6661 -2.5783 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6383 -1.9836 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6383 -1.9836 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9908 -1.4497 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9908 -1.4497 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2905 -1.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2905 -1.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3468 -2.3154 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3468 -2.3154 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.0840 -3.2013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0840 -3.2013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6661 -3.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6661 -3.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1716 -1.9763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1716 -1.9763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6337 -2.3808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6337 -2.3808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1593 -1.7621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1593 -1.7621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7773 -1.8888 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.7773 -1.8888 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.4061 -2.3788 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.4061 -2.3788 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.1284 -2.1709 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.1284 -2.1709 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.6442 -2.1653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.6442 -2.1653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2694 -1.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2694 -1.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1982 -2.0373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.1982 -2.0373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.3682 -1.8371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3682 -1.8371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7565 -2.8400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7565 -2.8400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4347 -2.6850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4347 -2.6850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9094 3.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9094 3.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3508 4.2319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3508 4.2319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1930 1.7765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1930 1.7765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1930 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1930 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8908 -1.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8908 -1.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6660 -1.2128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6660 -1.2128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 42 1 0 0 0 0 | + | 46 42 1 0 0 0 0 |
| − | 15 52 1 0 0 0 0 | + | 15 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 13 54 1 0 0 0 0 | + | 13 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 48 56 1 0 0 0 0 | + | 48 56 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 56 57 | + | M SAL 3 2 56 57 |
| − | M SBL 3 1 61 | + | M SBL 3 1 61 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 61 -0.5967 -1.2531 | + | M SVB 3 61 -0.5967 -1.2531 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 54 55 | + | M SAL 2 2 54 55 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 59 1.4025 1.5702 | + | M SVB 2 59 1.4025 1.5702 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 57 0.9094 3.3346 | + | M SVB 1 57 0.9094 3.3346 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAIGL0007 | + | ID FL7AAIGL0007 |
| − | KNApSAcK_ID C00006743 | + | KNApSAcK_ID C00006743 |
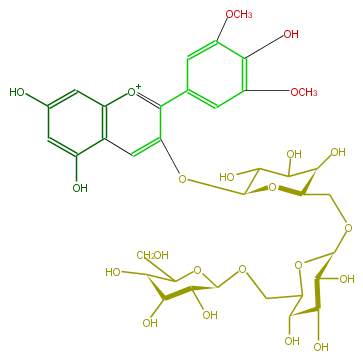
| − | NAME Malvidin 3-gentiotrioside | + | NAME Malvidin 3-gentiotrioside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C35H45O22 | + | FORMULA C35H45O22 |
| − | EXACTMASS 817.240248124 | + | EXACTMASS 817.240248124 |
| − | AVERAGEMASS 817.7186 | + | AVERAGEMASS 817.7186 |
| − | SMILES [C@H](OCC([C@@H]6O)O[C@H]([C@H]([C@@H](O)6)O)OC[C@@H](O2)[C@H](O)C(O)C([C@@H]2Oc(c3)c(c(c5)cc(OC)c(c(OC)5)O)[o+1]c(c4)c3c(cc(O)4)O)O)(O1)[C@H]([C@@H](O)[C@H](C1CO)O)O | + | SMILES [C@H](OCC([C@@H]6O)O[C@H]([C@H]([C@@H](O)6)O)OC[C@@H](O2)[C@H](O)C(O)C([C@@H]2Oc(c3)c(c(c5)cc(OC)c(c(OC)5)O)[o+1]c(c4)c3c(cc(O)4)O)O)(O1)[C@H]([C@@H](O)[C@H](C1CO)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
57 62 0 0 0 0 0 0 0 0999 V2000
-2.7223 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7223 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1660 0.4919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6097 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6097 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1660 1.7766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0534 0.4919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4971 0.8131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4971 1.4554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0534 1.7766 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.0590 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6260 1.4491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1930 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1930 2.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6260 2.7585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0590 2.4312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2784 1.7765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0590 2.9312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1660 -0.1502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0458 0.0314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4889 0.2274 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.1881 -0.2936 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7666 -0.1283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3486 -0.2936 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6494 0.2274 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0709 0.0622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9302 0.0777 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1125 0.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0038 0.5818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9001 -0.2936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2784 -0.9488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0840 -2.7285 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.6661 -2.5783 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6383 -1.9836 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9908 -1.4497 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2905 -1.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3468 -2.3154 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.0840 -3.2013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6661 -3.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1716 -1.9763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6337 -2.3808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1593 -1.7621 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7773 -1.8888 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.4061 -2.3788 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.1284 -2.1709 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.6442 -2.1653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2694 -1.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1982 -2.0373 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.3682 -1.8371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7565 -2.8400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4347 -2.6850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9094 3.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3508 4.2319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1930 1.7765 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1930 1.7765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8908 -1.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6660 -1.2128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
22 20 1 0 0 0 0
30 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
37 41 1 0 0 0 0
35 31 1 0 0 0 0
41 42 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 42 1 0 0 0 0
15 52 1 0 0 0 0
52 53 1 0 0 0 0
13 54 1 0 0 0 0
54 55 1 0 0 0 0
48 56 1 0 0 0 0
56 57 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 56 57
M SBL 3 1 61
M SMT 3 CH2OH
M SVB 3 61 -0.5967 -1.2531
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 54 55
M SBL 2 1 59
M SMT 2 OCH3
M SVB 2 59 1.4025 1.5702
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 57
M SMT 1 OCH3
M SVB 1 57 0.9094 3.3346
S SKP 8
ID FL7AAIGL0007
KNApSAcK_ID C00006743
NAME Malvidin 3-gentiotrioside
CAS_RN -
FORMULA C35H45O22
EXACTMASS 817.240248124
AVERAGEMASS 817.7186
SMILES [C@H](OCC([C@@H]6O)O[C@H]([C@H]([C@@H](O)6)O)OC[C@@H](O2)[C@H](O)C(O)C([C@@H]2Oc(c3)c(c(c5)cc(OC)c(c(OC)5)O)[o+1]c(c4)c3c(cc(O)4)O)O)(O1)[C@H]([C@@H](O)[C@H](C1CO)O)O
M END