Mol:FL7AACGL0114
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 86 93 0 0 0 0 0 0 0 0999 V2000 | + | 86 93 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.2822 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2822 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2822 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2822 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9967 1.2775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9967 1.2775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7112 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7112 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7112 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7112 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9967 2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9967 2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4257 1.2775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4257 1.2775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1401 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1401 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1401 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1401 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4257 2.9275 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 2.4257 2.9275 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9167 2.9633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9167 2.9633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6311 2.5508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6311 2.5508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3456 2.9633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3456 2.9633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3456 3.7883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3456 3.7883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6311 4.2008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6311 4.2008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9167 3.7883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9167 3.7883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9858 4.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9858 4.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7843 1.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7843 1.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2977 2.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2977 2.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9967 0.5148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9967 0.5148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9905 2.5910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9905 2.5910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.7005 -0.2829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.7005 -0.2829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.4687 0.0186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.4687 0.0186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.3808 0.8392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.3808 0.8392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.7233 1.5901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.7233 1.5901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.9551 1.2886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.9551 1.2886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.0429 0.4679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.0429 0.4679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4402 0.4000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4402 0.4000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.1063 -0.8327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1063 -0.8327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.1366 -0.3587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.1366 -0.3587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.9880 0.5743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.9880 0.5743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5308 -0.9629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5308 -0.9629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3556 -0.9905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3556 -0.9905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6002 -0.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6002 -0.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2123 0.3512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2123 0.3512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3875 0.3790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3875 0.3790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1428 -0.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1428 -0.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5626 -0.2327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5626 -0.2327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6855 -1.6285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6855 -1.6285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8985 -1.2344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8985 -1.2344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0527 -0.6862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0527 -0.6862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9687 -0.6114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9687 -0.6114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1405 -2.7991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1405 -2.7991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2786 -1.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2786 -1.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3558 -1.4612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3558 -1.4612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0510 -1.7004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0510 -1.7004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1891 -0.8881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1891 -0.8881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5547 -0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5547 -0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0557 -0.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0557 -0.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2752 -0.3649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2752 -0.3649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6566 0.1794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6566 0.1794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1131 -1.1727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1131 -1.1727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7316 -1.7169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7316 -1.7169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5696 -2.5247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5696 -2.5247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7879 -2.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7879 -2.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6257 -3.5975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6257 -3.5975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2451 -4.1425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2451 -4.1425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0267 -3.8786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0267 -3.8786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1890 -3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1890 -3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0830 -4.9503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0830 -4.9503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9451 -4.0506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9451 -4.0506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8023 -4.7626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8023 -4.7626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0258 2.8554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0258 2.8554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5790 2.1615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5790 2.1615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7917 2.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7917 2.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9882 2.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9882 2.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4349 2.9149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4349 2.9149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2223 2.6674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2223 2.6674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4074 3.2449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4074 3.2449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4850 2.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4850 2.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0143 2.4128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0143 2.4128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9320 1.7616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9320 1.7616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9746 3.5232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9746 3.5232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3865 4.2368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3865 4.2368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9746 4.9503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9746 4.9503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2104 4.2368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2104 4.2368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6223 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6223 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4462 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4462 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8587 4.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8587 4.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6837 4.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6837 4.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.0962 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.0962 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6837 2.8088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6837 2.8088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8587 2.8088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8587 2.8088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.9201 3.5232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.9201 3.5232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.2619 4.8159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.2619 4.8159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.9880 4.8159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.9880 4.8159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 42 38 1 0 0 0 0 | + | 42 38 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 42 47 1 0 0 0 0 | + | 42 47 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 50 52 1 0 0 0 0 | + | 50 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 2 0 0 0 0 | + | 54 55 2 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 2 0 0 0 0 | + | 56 57 2 0 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 58 59 2 0 0 0 0 | + | 58 59 2 0 0 0 0 |
| − | 59 54 1 0 0 0 0 | + | 59 54 1 0 0 0 0 |
| − | 57 60 1 0 0 0 0 | + | 57 60 1 0 0 0 0 |
| − | 56 61 1 0 0 0 0 | + | 56 61 1 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 49 28 1 0 0 0 0 | + | 49 28 1 0 0 0 0 |
| − | 63 64 1 1 0 0 0 | + | 63 64 1 1 0 0 0 |
| − | 64 65 1 1 0 0 0 | + | 64 65 1 1 0 0 0 |
| − | 66 65 1 1 0 0 0 | + | 66 65 1 1 0 0 0 |
| − | 66 67 1 0 0 0 0 | + | 66 67 1 0 0 0 0 |
| − | 67 68 1 0 0 0 0 | + | 67 68 1 0 0 0 0 |
| − | 68 63 1 0 0 0 0 | + | 68 63 1 0 0 0 0 |
| − | 68 69 1 0 0 0 0 | + | 68 69 1 0 0 0 0 |
| − | 63 70 1 0 0 0 0 | + | 63 70 1 0 0 0 0 |
| − | 64 71 1 0 0 0 0 | + | 64 71 1 0 0 0 0 |
| − | 65 72 1 0 0 0 0 | + | 65 72 1 0 0 0 0 |
| − | 73 74 1 0 0 0 0 | + | 73 74 1 0 0 0 0 |
| − | 74 75 2 0 0 0 0 | + | 74 75 2 0 0 0 0 |
| − | 74 76 1 0 0 0 0 | + | 74 76 1 0 0 0 0 |
| − | 76 77 2 0 0 0 0 | + | 76 77 2 0 0 0 0 |
| − | 77 78 1 0 0 0 0 | + | 77 78 1 0 0 0 0 |
| − | 78 79 2 0 0 0 0 | + | 78 79 2 0 0 0 0 |
| − | 79 80 1 0 0 0 0 | + | 79 80 1 0 0 0 0 |
| − | 80 81 2 0 0 0 0 | + | 80 81 2 0 0 0 0 |
| − | 81 82 1 0 0 0 0 | + | 81 82 1 0 0 0 0 |
| − | 82 83 2 0 0 0 0 | + | 82 83 2 0 0 0 0 |
| − | 83 78 1 0 0 0 0 | + | 83 78 1 0 0 0 0 |
| − | 81 84 1 0 0 0 0 | + | 81 84 1 0 0 0 0 |
| − | 80 85 1 0 0 0 0 | + | 80 85 1 0 0 0 0 |
| − | 85 86 1 0 0 0 0 | + | 85 86 1 0 0 0 0 |
| − | 73 69 1 0 0 0 0 | + | 73 69 1 0 0 0 0 |
| − | 19 66 1 0 0 0 0 | + | 19 66 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 35 18 1 0 0 0 0 | + | 35 18 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0114 | + | ID FL7AACGL0114 |
| − | KNApSAcK_ID C00014774 | + | KNApSAcK_ID C00014774 |
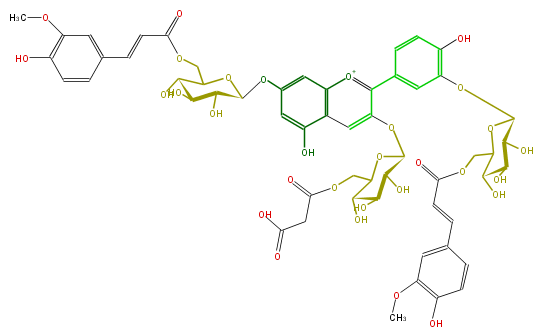
| − | NAME Cyanidin 3-(6-malonylglucoside)-7,3'-di-(6-feruloylglucoside) | + | NAME Cyanidin 3-(6-malonylglucoside)-7,3'-di-(6-feruloylglucoside) |
| − | CAS_RN 799854-19-6 | + | CAS_RN 799854-19-6 |
| − | FORMULA C56H59O30 | + | FORMULA C56H59O30 |
| − | EXACTMASS 1211.3091155480001 | + | EXACTMASS 1211.3091155480001 |
| − | AVERAGEMASS 1212.0496600000001 | + | AVERAGEMASS 1212.0496600000001 |
| − | SMILES c(c1)(C=CC(=O)OCC(C(O)8)OC(C(O)C8O)Oc(c2)c(O)ccc2c(c6OC(C7O)OC(C(C(O)7)O)COC(=O)CC(O)=O)[o+1]c(c3)c(c6)c(O)cc3OC(C(O)5)OC(C(C5O)O)COC(=O)C=Cc(c4)cc(OC)c(O)c4)ccc(O)c(OC)1 | + | SMILES c(c1)(C=CC(=O)OCC(C(O)8)OC(C(O)C8O)Oc(c2)c(O)ccc2c(c6OC(C7O)OC(C(C(O)7)O)COC(=O)CC(O)=O)[o+1]c(c3)c(c6)c(O)cc3OC(C(O)5)OC(C(C5O)O)COC(=O)C=Cc(c4)cc(OC)c(O)c4)ccc(O)c(OC)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
86 93 0 0 0 0 0 0 0 0999 V2000
0.2822 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2822 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9967 1.2775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7112 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7112 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9967 2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4257 1.2775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1401 1.6900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1401 2.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4257 2.9275 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
3.9167 2.9633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6311 2.5508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3456 2.9633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3456 3.7883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6311 4.2008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9167 3.7883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9858 4.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7843 1.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2977 2.8498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9967 0.5148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9905 2.5910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.7005 -0.2829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.4687 0.0186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.3808 0.8392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7233 1.5901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9551 1.2886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.0429 0.4679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4402 0.4000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.1063 -0.8327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.1366 -0.3587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.9880 0.5743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5308 -0.9629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3556 -0.9905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6002 -0.2023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2123 0.3512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3875 0.3790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1428 -0.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5626 -0.2327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6855 -1.6285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8985 -1.2344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0527 -0.6862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9687 -0.6114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1405 -2.7991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2786 -1.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3558 -1.4612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0510 -1.7004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1891 -0.8881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5547 -0.3624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0557 -0.1013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2752 -0.3649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6566 0.1794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1131 -1.1727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7316 -1.7169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5696 -2.5247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7879 -2.7886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6257 -3.5975 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2451 -4.1425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0267 -3.8786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1890 -3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0830 -4.9503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9451 -4.0506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8023 -4.7626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0258 2.8554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5790 2.1615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7917 2.4091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9882 2.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4349 2.9149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2223 2.6674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4074 3.2449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4850 2.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0143 2.4128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9320 1.7616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9746 3.5232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3865 4.2368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9746 4.9503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2104 4.2368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6223 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4462 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8587 4.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6837 4.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.0962 3.5232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6837 2.8088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8587 2.8088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.9201 3.5232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.2619 4.8159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.9880 4.8159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
8 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
13 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
34 41 1 0 0 0 0
42 38 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
44 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
42 47 1 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
50 52 1 0 0 0 0
52 53 2 0 0 0 0
53 54 1 0 0 0 0
54 55 2 0 0 0 0
55 56 1 0 0 0 0
56 57 2 0 0 0 0
57 58 1 0 0 0 0
58 59 2 0 0 0 0
59 54 1 0 0 0 0
57 60 1 0 0 0 0
56 61 1 0 0 0 0
61 62 1 0 0 0 0
49 28 1 0 0 0 0
63 64 1 1 0 0 0
64 65 1 1 0 0 0
66 65 1 1 0 0 0
66 67 1 0 0 0 0
67 68 1 0 0 0 0
68 63 1 0 0 0 0
68 69 1 0 0 0 0
63 70 1 0 0 0 0
64 71 1 0 0 0 0
65 72 1 0 0 0 0
73 74 1 0 0 0 0
74 75 2 0 0 0 0
74 76 1 0 0 0 0
76 77 2 0 0 0 0
77 78 1 0 0 0 0
78 79 2 0 0 0 0
79 80 1 0 0 0 0
80 81 2 0 0 0 0
81 82 1 0 0 0 0
82 83 2 0 0 0 0
83 78 1 0 0 0 0
81 84 1 0 0 0 0
80 85 1 0 0 0 0
85 86 1 0 0 0 0
73 69 1 0 0 0 0
19 66 1 0 0 0 0
25 21 1 0 0 0 0
35 18 1 0 0 0 0
S SKP 8
ID FL7AACGL0114
KNApSAcK_ID C00014774
NAME Cyanidin 3-(6-malonylglucoside)-7,3'-di-(6-feruloylglucoside)
CAS_RN 799854-19-6
FORMULA C56H59O30
EXACTMASS 1211.3091155480001
AVERAGEMASS 1212.0496600000001
SMILES c(c1)(C=CC(=O)OCC(C(O)8)OC(C(O)C8O)Oc(c2)c(O)ccc2c(c6OC(C7O)OC(C(C(O)7)O)COC(=O)CC(O)=O)[o+1]c(c3)c(c6)c(O)cc3OC(C(O)5)OC(C(C5O)O)COC(=O)C=Cc(c4)cc(OC)c(O)c4)ccc(O)c(OC)1
M END