Mol:FL7AACGL0051
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0564 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0564 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0564 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0564 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5001 -1.1695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5001 -1.1695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0562 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0562 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0562 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0562 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5001 0.1152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5001 0.1152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6125 -1.1695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6125 -1.1695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1688 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1688 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1688 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1688 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6125 0.1152 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.6125 0.1152 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7249 0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7249 0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2919 -0.2122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2919 -0.2122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8589 0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8589 0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8589 0.7698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8589 0.7698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2919 1.0972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2919 1.0972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7249 0.7698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7249 0.7698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6125 0.1151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6125 0.1151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4257 1.0971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4257 1.0971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5001 -1.8116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5001 -1.8116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5849 -1.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5849 -1.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2919 1.7517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2919 1.7517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6589 -2.0858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6589 -2.0858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2877 -2.5758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2877 -2.5758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7532 -2.3679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7532 -2.3679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2374 -2.3623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2374 -2.3623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6122 -1.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6122 -1.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0798 -2.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0798 -2.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1727 -2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1727 -2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8663 -2.6039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8663 -2.6039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4469 -2.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4469 -2.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5447 -1.7322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5447 -1.7322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8454 -1.2277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8454 -1.2277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5446 -1.7487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5446 -1.7487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1231 -1.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1231 -1.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7051 -1.7487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7051 -1.7487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0060 -1.2277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0060 -1.2277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4275 -1.3930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4275 -1.3930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2868 -1.3774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2868 -1.3774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6656 -0.9804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6656 -0.9804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7093 -1.4161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7093 -1.4161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5447 -1.0309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5447 -1.0309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1944 -0.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1944 -0.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7197 -0.9424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7197 -0.9424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1872 0.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1872 0.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6514 0.3866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6514 0.3866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6514 0.9214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6514 0.9214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0853 1.2483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0853 1.2483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0853 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0853 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6514 2.2289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6514 2.2289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2176 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2176 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2176 1.2483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2176 1.2483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6514 2.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6514 2.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7832 2.2286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7832 2.2286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0687 -1.7618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0687 -1.7618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7832 -2.1743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7832 -2.1743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 31 41 1 0 0 0 0 | + | 31 41 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 41 1 0 0 0 0 | + | 42 41 1 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 49 52 1 0 0 0 0 | + | 49 52 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 35 54 1 0 0 0 0 | + | 35 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 59 -4.8549 3.4522 | + | M SBV 1 59 -4.8549 3.4522 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0051 | + | ID FL7AACGL0051 |
| − | KNApSAcK_ID C00006822 | + | KNApSAcK_ID C00006822 |
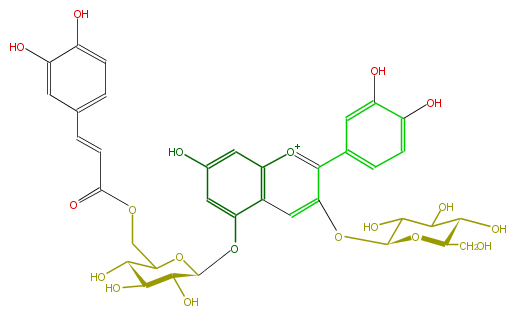
| − | NAME Gentiocyanin A;Cyanidin 3-glucoside-5-(6-caffeoylglucoside) | + | NAME Gentiocyanin A;Cyanidin 3-glucoside-5-(6-caffeoylglucoside) |
| − | CAS_RN 170663-59-9 | + | CAS_RN 170663-59-9 |
| − | FORMULA C36H37O19 | + | FORMULA C36H37O19 |
| − | EXACTMASS 773.192904002 | + | EXACTMASS 773.192904002 |
| − | AVERAGEMASS 773.66758 | + | AVERAGEMASS 773.66758 |
| − | SMILES C(COC(C=Cc(c6)ccc(c(O)6)O)=O)(C(O)1)OC(Oc(c2)c(c4)c([o+1]c(c(OC(O5)C(C(O)C(C(CO)5)O)O)4)c(c3)ccc(O)c(O)3)cc2O)C(O)C(O)1 | + | SMILES C(COC(C=Cc(c6)ccc(c(O)6)O)=O)(C(O)1)OC(Oc(c2)c(c4)c([o+1]c(c(OC(O5)C(C(O)C(C(CO)5)O)O)4)c(c3)ccc(O)c(O)3)cc2O)C(O)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.0564 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0564 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5001 -1.1695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0562 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0562 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5001 0.1152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6125 -1.1695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1688 -0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1688 -0.2059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6125 0.1152 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.7249 0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2919 -0.2122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8589 0.1151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8589 0.7698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2919 1.0972 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7249 0.7698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6125 0.1151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4257 1.0971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5001 -1.8116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5849 -1.5690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2919 1.7517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6589 -2.0858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2877 -2.5758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7532 -2.3679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2374 -2.3623 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6122 -1.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0798 -2.2343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1727 -2.3825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8663 -2.6039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4469 -2.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5447 -1.7322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8454 -1.2277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5446 -1.7487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1231 -1.5834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7051 -1.7487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0060 -1.2277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4275 -1.3930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2868 -1.3774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6656 -0.9804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7093 -1.4161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5447 -1.0309 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1944 -0.6391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7197 -0.9424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1872 0.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6514 0.3866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6514 0.9214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0853 1.2483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0853 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6514 2.2289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2176 1.9020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2176 1.2483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6514 2.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7832 2.2286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0687 -1.7618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7832 -2.1743 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 31 1 0 0 0 0
25 19 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
33 20 1 0 0 0 0
31 41 1 0 0 0 0
42 43 2 0 0 0 0
42 41 1 0 0 0 0
42 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
49 52 1 0 0 0 0
50 53 1 0 0 0 0
35 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 59
M SMT 1 CH2OH
M SBV 1 59 -4.8549 3.4522
S SKP 8
ID FL7AACGL0051
KNApSAcK_ID C00006822
NAME Gentiocyanin A;Cyanidin 3-glucoside-5-(6-caffeoylglucoside)
CAS_RN 170663-59-9
FORMULA C36H37O19
EXACTMASS 773.192904002
AVERAGEMASS 773.66758
SMILES C(COC(C=Cc(c6)ccc(c(O)6)O)=O)(C(O)1)OC(Oc(c2)c(c4)c([o+1]c(c(OC(O5)C(C(O)C(C(CO)5)O)O)4)c(c3)ccc(O)c(O)3)cc2O)C(O)C(O)1
M END