Mol:FL7AACGL0048
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0414 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0414 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0414 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0414 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4851 -0.2978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4851 -0.2978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9288 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9288 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9288 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9288 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4851 0.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4851 0.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3725 -0.2978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3725 -0.2978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1838 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1838 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1838 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1838 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3725 0.9869 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.3725 0.9869 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7399 0.9868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7399 0.9868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3069 0.6594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3069 0.6594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8739 0.9868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8739 0.9868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8739 1.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8739 1.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3069 1.9688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3069 1.9688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7399 1.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7399 1.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5975 0.9868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5975 0.9868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4407 1.9687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4407 1.9687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4851 -0.9400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4851 -0.9400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5999 -0.6974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5999 -0.6974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3069 2.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3069 2.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6438 -1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6438 -1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2727 -1.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2727 -1.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7382 -1.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7382 -1.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2224 -1.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2224 -1.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5972 -1.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5972 -1.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0648 -1.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0648 -1.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1577 -1.5108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1577 -1.5108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8513 -1.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8513 -1.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4319 -2.0104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4319 -2.0104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5297 -0.8605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5297 -0.8605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8605 -0.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8605 -0.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5597 -0.8771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5597 -0.8771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1381 -0.7117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1381 -0.7117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7201 -0.8771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7201 -0.8771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0210 -0.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0210 -0.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4425 -0.5213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4425 -0.5213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3018 -0.5057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3018 -0.5057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6807 -0.1088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6807 -0.1088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7243 -0.5445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7243 -0.5445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2869 -0.8812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2869 -0.8812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5297 -0.1592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5297 -0.1592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7613 -1.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7613 -1.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5825 -2.3565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5825 -2.3565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8516 -2.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8516 -2.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4825 -1.8505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4825 -1.8505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7999 -1.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7999 -1.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4690 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4690 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8074 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8074 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4765 -1.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4765 -1.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8074 -2.4408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8074 -2.4408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4690 -2.4408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4690 -2.4408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1849 -1.8677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1849 -1.8677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1577 -2.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1577 -2.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 31 42 1 0 0 0 0 | + | 31 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 44 54 2 0 0 0 0 | + | 44 54 2 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0048 | + | ID FL7AACGL0048 |
| − | KNApSAcK_ID C00006819 | + | KNApSAcK_ID C00006819 |
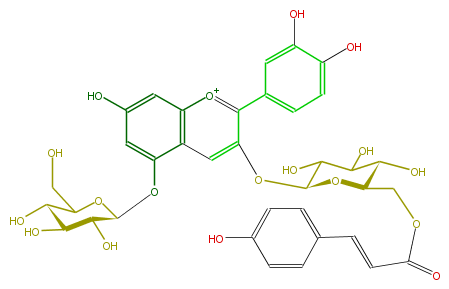
| − | NAME Perillanin;Cyanidin 3-(6-p-coumaroylglucoside)-5-glucoside | + | NAME Perillanin;Cyanidin 3-(6-p-coumaroylglucoside)-5-glucoside |
| − | CAS_RN 152885-69-3 | + | CAS_RN 152885-69-3 |
| − | FORMULA C36H37O18 | + | FORMULA C36H37O18 |
| − | EXACTMASS 757.1979893800001 | + | EXACTMASS 757.1979893800001 |
| − | AVERAGEMASS 757.66818 | + | AVERAGEMASS 757.66818 |
| − | SMILES c(c1)(c([o+1]6)c(cc(c46)c(OC(O5)C(O)C(O)C(O)C5CO)cc(O)c4)OC(C(O)3)OC(C(C3O)O)COC(=O)C=Cc(c2)ccc(c2)O)ccc(c(O)1)O | + | SMILES c(c1)(c([o+1]6)c(cc(c46)c(OC(O5)C(O)C(O)C(O)C5CO)cc(O)c4)OC(C(O)3)OC(C(C3O)O)COC(=O)C=Cc(c2)ccc(c2)O)ccc(c(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.0414 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0414 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4851 -0.2978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9288 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9288 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4851 0.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3725 -0.2978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1838 0.0233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1838 0.6657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3725 0.9869 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.7399 0.9868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3069 0.6594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8739 0.9868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8739 1.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3069 1.9688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7399 1.6415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5975 0.9868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4407 1.9687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4851 -0.9400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5999 -0.6974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3069 2.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6438 -1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2727 -1.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7382 -1.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2224 -1.4907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5972 -1.1158 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0648 -1.3626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1577 -1.5108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8513 -1.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4319 -2.0104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5297 -0.8605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8605 -0.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5597 -0.8771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1381 -0.7117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7201 -0.8771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0210 -0.3560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4425 -0.5213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3018 -0.5057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6807 -0.1088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7243 -0.5445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2869 -0.8812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5297 -0.1592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7613 -1.5824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5825 -2.3565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8516 -2.4678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4825 -1.8505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7999 -1.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4690 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8074 -1.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4765 -1.8677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8074 -2.4408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4690 -2.4408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1849 -1.8677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1577 -2.6233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 31 1 0 0 0 0
25 19 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
35 41 1 0 0 0 0
33 20 1 0 0 0 0
31 42 1 0 0 0 0
41 43 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
44 45 1 0 0 0 0
44 54 2 0 0 0 0
43 44 1 0 0 0 0
S SKP 8
ID FL7AACGL0048
KNApSAcK_ID C00006819
NAME Perillanin;Cyanidin 3-(6-p-coumaroylglucoside)-5-glucoside
CAS_RN 152885-69-3
FORMULA C36H37O18
EXACTMASS 757.1979893800001
AVERAGEMASS 757.66818
SMILES c(c1)(c([o+1]6)c(cc(c46)c(OC(O5)C(O)C(O)C(O)C5CO)cc(O)c4)OC(C(O)3)OC(C(C3O)O)COC(=O)C=Cc(c2)ccc(c2)O)ccc(c(O)1)O
M END