Mol:FL7AACGL0035
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.2923 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2923 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2923 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2923 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7360 1.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7360 1.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1797 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1797 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1797 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1797 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7360 2.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7360 2.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6234 1.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6234 1.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0671 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0671 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0671 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0671 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6234 2.9554 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.6234 2.9554 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4890 2.9553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4890 2.9553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0560 2.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0560 2.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6230 2.9553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6230 2.9553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6230 3.6099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6230 3.6099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0560 3.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0560 3.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4890 3.6099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4890 3.6099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8484 2.9553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8484 2.9553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1898 3.9372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1898 3.9372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7360 1.0285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7360 1.0285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3490 1.2711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3490 1.2711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8275 -0.0430 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.8275 -0.0430 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.3205 0.3447 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.3205 0.3447 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4516 -0.3364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4516 -0.3364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2769 -0.9514 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.2769 -0.9514 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.8275 -1.2693 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.8275 -1.2693 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.6529 -0.6580 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.6529 -0.6580 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.0023 0.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0023 0.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1543 -0.7019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1543 -0.7019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1511 -2.2155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1511 -2.2155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2418 -1.8064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2418 -1.8064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6720 0.4594 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.6720 0.4594 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.9755 -0.1326 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9755 -0.1326 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5842 -0.0115 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.5842 -0.0115 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.1349 -0.1025 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.1349 -0.1025 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8059 0.3675 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.8059 0.3675 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2611 0.1923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2611 0.1923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3535 -0.0539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3535 -0.0539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8140 -0.6170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8140 -0.6170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0882 -0.4047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0882 -0.4047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2541 -2.7749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2541 -2.7749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0808 -3.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0808 -3.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0954 -4.0315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0954 -4.0315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6991 -3.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6991 -3.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9467 -3.9981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9467 -3.9981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6259 -4.0105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6259 -4.0105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9523 -3.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9523 -3.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5823 -3.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5823 -3.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8859 -4.0334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8859 -4.0334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5595 -4.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5595 -4.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9295 -4.5803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9295 -4.5803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5148 -4.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5148 -4.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0560 4.5918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0560 4.5918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6648 0.8796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6648 0.8796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5378 0.3919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5378 0.3919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 48 51 1 0 0 0 0 | + | 48 51 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 15 52 1 0 0 0 0 | + | 15 52 1 0 0 0 0 |
| − | 35 53 1 0 0 0 0 | + | 35 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 58 3.5148 0.392 | + | M SVB 1 58 3.5148 0.392 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0035 | + | ID FL7AACGL0035 |
| − | KNApSAcK_ID C00006805 | + | KNApSAcK_ID C00006805 |
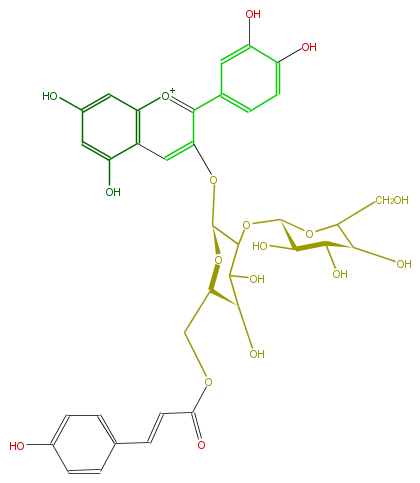
| − | NAME Cyanidin 3-(6''-p-coumarylsophoroside) | + | NAME Cyanidin 3-(6''-p-coumarylsophoroside) |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C36H37O18 | + | FORMULA C36H37O18 |
| − | EXACTMASS 757.1979893800001 | + | EXACTMASS 757.1979893800001 |
| − | AVERAGEMASS 757.66818 | + | AVERAGEMASS 757.66818 |
| − | SMILES c(c6)(O)cc(c(c65)cc(c([o+1]5)c(c4)cc(O)c(c4)O)O[C@H](O1)C(O[C@H]([C@H](O)3)OC(CO)[C@H]([C@@H](O)3)O)C([C@@H](O)[C@H]1COC(C=Cc(c2)ccc(c2)O)=O)O)O | + | SMILES c(c6)(O)cc(c(c65)cc(c([o+1]5)c(c4)cc(O)c(c4)O)O[C@H](O1)C(O[C@H]([C@H](O)3)OC(CO)[C@H]([C@@H](O)3)O)C([C@@H](O)[C@H]1COC(C=Cc(c2)ccc(c2)O)=O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.2923 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2923 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7360 1.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1797 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1797 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7360 2.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6234 1.6706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0671 1.9918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0671 2.6342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6234 2.9554 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.4890 2.9553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0560 2.6279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6230 2.9553 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6230 3.6099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0560 3.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4890 3.6099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8484 2.9553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1898 3.9372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7360 1.0285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3490 1.2711 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8275 -0.0430 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.3205 0.3447 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4516 -0.3364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2769 -0.9514 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8275 -1.2693 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.6529 -0.6580 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.0023 0.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1543 -0.7019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1511 -2.2155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2418 -1.8064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6720 0.4594 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.9755 -0.1326 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5842 -0.0115 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.1349 -0.1025 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8059 0.3675 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2611 0.1923 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3535 -0.0539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8140 -0.6170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0882 -0.4047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2541 -2.7749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0808 -3.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0954 -4.0315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6991 -3.4160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9467 -3.9981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6259 -4.0105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9523 -3.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5823 -3.4636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8859 -4.0334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5595 -4.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9295 -4.5803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5148 -4.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0560 4.5918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6648 0.8796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5378 0.3919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
27 31 1 0 0 0 0
22 20 1 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
40 30 1 0 0 0 0
15 52 1 0 0 0 0
35 53 1 0 0 0 0
53 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 58
M SMT 1 CH2OH
M SVB 1 58 3.5148 0.392
S SKP 8
ID FL7AACGL0035
KNApSAcK_ID C00006805
NAME Cyanidin 3-(6''-p-coumarylsophoroside)
CAS_RN -
FORMULA C36H37O18
EXACTMASS 757.1979893800001
AVERAGEMASS 757.66818
SMILES c(c6)(O)cc(c(c65)cc(c([o+1]5)c(c4)cc(O)c(c4)O)O[C@H](O1)C(O[C@H]([C@H](O)3)OC(CO)[C@H]([C@@H](O)3)O)C([C@@H](O)[C@H]1COC(C=Cc(c2)ccc(c2)O)=O)O)O
M END