Mol:FL7AAAGL0073
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6949 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6949 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6949 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6949 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9804 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9804 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2660 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2660 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2660 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2660 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9804 2.8478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9804 2.8478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4485 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4485 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1630 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1630 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1630 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1630 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4485 2.8478 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.4485 2.8478 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9395 2.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9395 2.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6540 2.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6540 2.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3685 2.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3685 2.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3685 3.7086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3685 3.7086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6540 4.1211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6540 4.1211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9395 3.7086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9395 3.7086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0087 4.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0087 4.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2749 2.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2749 2.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9804 0.4351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9804 0.4351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8132 1.2349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8132 1.2349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5255 0.4839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5255 0.4839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1128 -0.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1128 -0.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3145 -0.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3145 -0.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5210 -0.2485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5210 -0.2485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9336 0.4662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9336 0.4662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7320 0.2571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7320 0.2571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8889 0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8889 0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0087 0.0007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0087 0.0007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5615 0.0282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5615 0.0282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4860 -0.6616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4860 -0.6616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7245 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7245 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4197 -1.0246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4197 -1.0246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0323 -0.4716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0323 -0.4716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8412 -0.3081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8412 -0.3081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1461 0.1368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1461 0.1368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5334 -0.4161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5334 -0.4161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4905 -0.9736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4905 -0.9736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4914 -1.3142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4914 -1.3142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9879 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9879 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0569 0.1059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0569 0.1059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4510 1.1353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4510 1.1353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4241 -0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4241 -0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0719 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0719 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0628 -1.6152 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0628 -1.6152 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7900 -0.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7900 -0.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4988 -0.8236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4988 -0.8236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4888 -1.6474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4888 -1.6474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1982 -2.0686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1982 -2.0686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1881 -2.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1881 -2.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4687 -3.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4687 -3.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7593 -2.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7593 -2.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7693 -2.0512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7693 -2.0512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4586 -4.1211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4586 -4.1211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 41 27 1 0 0 0 0 | + | 41 27 1 0 0 0 0 |
| − | 42 40 1 0 0 0 0 | + | 42 40 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0073 | + | ID FL7AAAGL0073 |
| − | KNApSAcK_ID C00014852 | + | KNApSAcK_ID C00014852 |
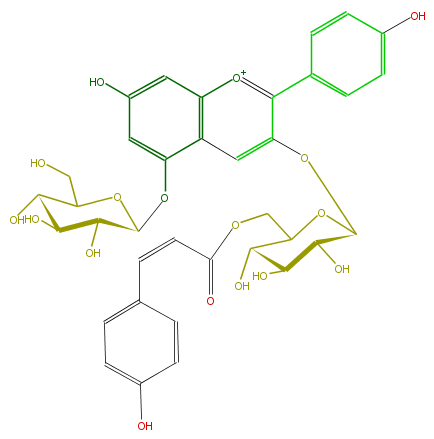
| − | NAME Pelargonidin 3-[6-((Z)-p-coumaroyl)glucoside]-5-glucoside | + | NAME Pelargonidin 3-[6-((Z)-p-coumaroyl)glucoside]-5-glucoside |
| − | CAS_RN 167936-44-9 | + | CAS_RN 167936-44-9 |
| − | FORMULA C36H37O17 | + | FORMULA C36H37O17 |
| − | EXACTMASS 741.203074758 | + | EXACTMASS 741.203074758 |
| − | AVERAGEMASS 741.66878 | + | AVERAGEMASS 741.66878 |
| − | SMILES OC(C1O)C(CO)OC(Oc(c24)cc(cc([o+1]c(c(OC(O5)C(O)C(C(O)C5COC(=O)C=Cc(c6)ccc(c6)O)O)c4)c(c3)ccc(c3)O)2)O)C1O | + | SMILES OC(C1O)C(CO)OC(Oc(c24)cc(cc([o+1]c(c(OC(O5)C(O)C(C(O)C5COC(=O)C=Cc(c6)ccc(c6)O)O)c4)c(c3)ccc(c3)O)2)O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.6949 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6949 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9804 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2660 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2660 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9804 2.8478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4485 1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1630 1.6103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1630 2.4353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4485 2.8478 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.9395 2.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6540 2.4711 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3685 2.8836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3685 3.7086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6540 4.1211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9395 3.7086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0087 4.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2749 2.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9804 0.4351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8132 1.2349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5255 0.4839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1128 -0.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3145 -0.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5210 -0.2485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9336 0.4662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7320 0.2571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8889 0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0087 0.0007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5615 0.0282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4860 -0.6616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7245 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4197 -1.0246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0323 -0.4716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8412 -0.3081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1461 0.1368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5334 -0.4161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4905 -0.9736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4914 -1.3142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9879 -1.1194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0569 0.1059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4510 1.1353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4241 -0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0719 -0.8075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0628 -1.6152 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7900 -0.4036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4988 -0.8236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4888 -1.6474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1982 -2.0686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1881 -2.8936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4687 -3.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7593 -2.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7693 -2.0512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4586 -4.1211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
33 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
36 40 1 0 0 0 0
34 20 1 0 0 0 0
41 27 1 0 0 0 0
42 40 1 0 0 0 0
24 19 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
42 43 1 0 0 0 0
S SKP 8
ID FL7AAAGL0073
KNApSAcK_ID C00014852
NAME Pelargonidin 3-[6-((Z)-p-coumaroyl)glucoside]-5-glucoside
CAS_RN 167936-44-9
FORMULA C36H37O17
EXACTMASS 741.203074758
AVERAGEMASS 741.66878
SMILES OC(C1O)C(CO)OC(Oc(c24)cc(cc([o+1]c(c(OC(O5)C(O)C(C(O)C5COC(=O)C=Cc(c6)ccc(c6)O)O)c4)c(c3)ccc(c3)O)2)O)C1O
M END