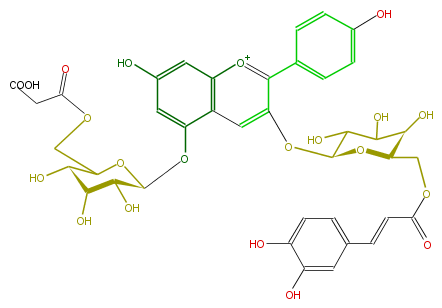
Mol:FL7AAAGL0036
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 60 65 0 0 0 0 0 0 0 0999 V2000 | + | 60 65 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8224 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8224 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8224 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8224 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2661 0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2661 0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2902 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2902 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2902 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2902 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2661 1.8233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2661 1.8233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8465 0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8465 0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4028 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4028 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4028 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4028 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8465 1.8233 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.8465 1.8233 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9589 1.8232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9589 1.8232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5259 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5259 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0929 1.8232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0929 1.8232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0929 2.4779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0929 2.4779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5259 2.8052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5259 2.8052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9589 2.4779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9589 2.4779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3785 1.8232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3785 1.8232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6597 2.8051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6597 2.8051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2661 -0.1035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2661 -0.1035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8189 0.1391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8189 0.1391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9764 0.4391 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.9764 0.4391 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.6755 -0.0819 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6755 -0.0819 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2540 0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2540 0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8360 -0.0819 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.8360 -0.0819 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.1369 0.4391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.1369 0.4391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5584 0.2739 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.5584 0.2739 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.4177 0.2894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4177 0.2894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6000 0.7484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6000 0.7484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4913 0.7935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4913 0.7935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3876 -0.0819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3876 -0.0819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5898 -0.2746 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.5898 -0.2746 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.2187 -0.7646 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.2187 -0.7646 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6842 -0.5567 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6842 -0.5567 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0732 -0.7048 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.0732 -0.7048 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5432 -0.1763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5432 -0.1763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0108 -0.4231 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.0108 -0.4231 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.1473 -0.4775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1473 -0.4775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3414 -1.3306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3414 -1.3306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3779 -1.0708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3779 -1.0708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8685 0.0910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8685 0.0910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5781 -0.7931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5781 -0.7931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2887 -1.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2887 -1.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7736 -1.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7736 -1.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4792 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4792 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9899 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9899 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7007 -1.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7007 -1.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1223 -1.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1223 -1.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8331 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8331 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1223 -2.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1223 -2.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7007 -2.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7007 -2.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2556 -1.8042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2556 -1.8042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5844 -1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5844 -1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1996 0.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1996 0.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7453 1.1852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7453 1.1852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2420 1.0044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2420 1.0044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6954 1.3848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6954 1.3848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3471 1.1477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3471 1.1477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6528 1.7096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6528 1.7096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5750 2.0673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5750 2.0673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8336 -2.8052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8336 -2.8052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 30 41 1 0 0 0 0 | + | 30 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 48 51 1 0 0 0 0 | + | 48 51 1 0 0 0 0 |
| − | 42 52 2 0 0 0 0 | + | 42 52 2 0 0 0 0 |
| − | 40 53 1 0 0 0 0 | + | 40 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 2 0 0 0 0 | + | 56 57 2 0 0 0 0 |
| − | 54 58 2 0 0 0 0 | + | 54 58 2 0 0 0 0 |
| − | 56 59 1 0 0 0 0 | + | 56 59 1 0 0 0 0 |
| − | 49 60 1 0 0 0 0 | + | 49 60 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 56 57 59 | + | M SAL 1 3 56 57 59 |
| − | M SBL 1 1 61 | + | M SBL 1 1 61 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 61 -3.9837 1.0648 | + | M SVB 1 61 -3.9837 1.0648 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0036 | + | ID FL7AAAGL0036 |
| − | KNApSAcK_ID C00006775 | + | KNApSAcK_ID C00006775 |
| − | NAME Monodemalonylsalvianin | + | NAME Monodemalonylsalvianin |
| − | CAS_RN 128508-43-0 | + | CAS_RN 128508-43-0 |
| − | FORMULA C39H39O21 | + | FORMULA C39H39O21 |
| − | EXACTMASS 843.19838331 | + | EXACTMASS 843.19838331 |
| − | AVERAGEMASS 843.7143599999999 | + | AVERAGEMASS 843.7143599999999 |
| − | SMILES c(c64)(c(cc(c6)O)O[C@@H]([C@H]5O)OC([C@@H]([C@@H]5O)O)COC(CC(O)=O)=O)cc(c([o+1]4)c(c3)ccc(O)c3)O[C@@H](C2O)O[C@@H]([C@H](O)C2O)COC(=O)C=Cc(c1)ccc(O)c1O | + | SMILES c(c64)(c(cc(c6)O)O[C@@H]([C@H]5O)OC([C@@H]([C@@H]5O)O)COC(CC(O)=O)=O)cc(c([o+1]4)c(c3)ccc(O)c3)O[C@@H](C2O)O[C@@H]([C@H](O)C2O)COC(=O)C=Cc(c1)ccc(O)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
60 65 0 0 0 0 0 0 0 0999 V2000
-0.8224 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8224 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2661 0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2902 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2902 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2661 1.8233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8465 0.5386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4028 0.8598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4028 1.5021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8465 1.8233 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.9589 1.8232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5259 1.4958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0929 1.8232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0929 2.4779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5259 2.8052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9589 2.4779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3785 1.8232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6597 2.8051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2661 -0.1035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8189 0.1391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9764 0.4391 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.6755 -0.0819 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2540 0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8360 -0.0819 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.1369 0.4391 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5584 0.2739 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4177 0.2894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6000 0.7484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4913 0.7935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3876 -0.0819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5898 -0.2746 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.2187 -0.7646 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6842 -0.5567 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0732 -0.7048 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5432 -0.1763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0108 -0.4231 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.1473 -0.4775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3414 -1.3306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3779 -1.0708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8685 0.0910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5781 -0.7931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2887 -1.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7736 -1.2944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4792 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9899 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7007 -1.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1223 -1.3033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8331 -1.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1223 -2.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7007 -2.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2556 -1.8042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5844 -1.8064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1996 0.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7453 1.1852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2420 1.0044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6954 1.3848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3471 1.1477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6528 1.7096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5750 2.0673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8336 -2.8052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
22 20 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
34 19 1 0 0 0 0
30 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
42 52 2 0 0 0 0
40 53 1 0 0 0 0
53 54 1 0 0 0 0
54 55 1 0 0 0 0
55 56 1 0 0 0 0
56 57 2 0 0 0 0
54 58 2 0 0 0 0
56 59 1 0 0 0 0
49 60 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 56 57 59
M SBL 1 1 61
M SMT 1 COOH
M SVB 1 61 -3.9837 1.0648
S SKP 8
ID FL7AAAGL0036
KNApSAcK_ID C00006775
NAME Monodemalonylsalvianin
CAS_RN 128508-43-0
FORMULA C39H39O21
EXACTMASS 843.19838331
AVERAGEMASS 843.7143599999999
SMILES c(c64)(c(cc(c6)O)O[C@@H]([C@H]5O)OC([C@@H]([C@@H]5O)O)COC(CC(O)=O)=O)cc(c([o+1]4)c(c3)ccc(O)c3)O[C@@H](C2O)O[C@@H]([C@H](O)C2O)COC(=O)C=Cc(c1)ccc(O)c1O
M END