Mol:FL7AAAGL0025
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3366 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3366 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3366 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3366 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7803 -0.3286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7803 -0.3286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2240 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2240 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2240 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2240 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7803 0.9561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7803 0.9561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3323 -0.3286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3323 -0.3286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8886 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8886 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8886 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8886 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3323 0.9561 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.3323 0.9561 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4447 0.9560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4447 0.9560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0117 0.6286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0117 0.6286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5786 0.9560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5786 0.9560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5786 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5786 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0117 1.9380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0117 1.9380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4447 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4447 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8927 0.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8927 0.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1454 1.9379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1454 1.9379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7803 -0.9707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7803 -0.9707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3047 -0.7281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3047 -0.7281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4621 -0.4281 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4621 -0.4281 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.1613 -0.9491 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1613 -0.9491 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7398 -0.7838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7398 -0.7838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3217 -0.9491 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.3217 -0.9491 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.6226 -0.4281 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.6226 -0.4281 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0441 -0.5933 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.0441 -0.5933 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.9034 -0.5778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9034 -0.5778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0857 -0.1188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0857 -0.1188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0567 -0.6787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0567 -0.6787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1041 -1.1418 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.1041 -1.1418 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.7329 -1.6318 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7329 -1.6318 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1984 -1.4239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1984 -1.4239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5874 -1.5719 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5874 -1.5719 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.0574 -1.0435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0574 -1.0435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5250 -1.2903 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.5250 -1.2903 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.6615 -1.3447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6615 -1.3447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8556 -2.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8556 -2.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8922 -1.9380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8922 -1.9380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3828 -0.7763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3828 -0.7763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7244 -0.1992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7244 -0.1992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1528 0.1602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1528 0.1602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0400 0.7997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0400 0.7997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5396 1.2188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5396 1.2188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4264 1.8607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4264 1.8607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0169 1.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0169 1.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5381 0.0924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5381 0.0924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4922 -1.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4922 -1.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4922 -2.0956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4922 -2.0956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 33 19 1 0 0 0 0 | + | 33 19 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 41 46 2 0 0 0 0 | + | 41 46 2 0 0 0 0 |
| − | 24 47 1 0 0 0 0 | + | 24 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 47 48 | + | M SAL 2 2 47 48 |
| − | M SBL 2 1 51 | + | M SBL 2 1 51 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 51 3.6854 -0.9622 | + | M SVB 2 51 3.6854 -0.9622 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 43 44 45 | + | M SAL 1 3 43 44 45 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 47 -3.9599 1.0045 | + | M SVB 1 47 -3.9599 1.0045 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0025 | + | ID FL7AAAGL0025 |
| − | KNApSAcK_ID C00006764 | + | KNApSAcK_ID C00006764 |
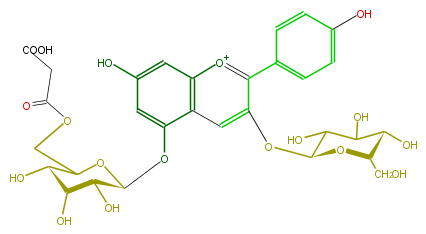
| − | NAME Pelargonidin 3-glucoside-5-(6'''-malonylglucoside) | + | NAME Pelargonidin 3-glucoside-5-(6'''-malonylglucoside) |
| − | CAS_RN 167774-88-1 | + | CAS_RN 167774-88-1 |
| − | FORMULA C30H33O18 | + | FORMULA C30H33O18 |
| − | EXACTMASS 681.166689252 | + | EXACTMASS 681.166689252 |
| − | AVERAGEMASS 681.57222 | + | AVERAGEMASS 681.57222 |
| − | SMILES c(c21)c(O[C@@H](C(O)5)O[C@@H]([C@@H](C5O)O)CO)c(c(c4)ccc(O)c4)[o+1]c1cc(cc(O[C@H](O3)[C@@H](O)[C@H]([C@@H](O)C3COC(CC(O)=O)=O)O)2)O | + | SMILES c(c21)c(O[C@@H](C(O)5)O[C@@H]([C@@H](C5O)O)CO)c(c(c4)ccc(O)c4)[o+1]c1cc(cc(O[C@H](O3)[C@@H](O)[C@H]([C@@H](O)C3COC(CC(O)=O)=O)O)2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-1.3366 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3366 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7803 -0.3286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2240 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2240 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7803 0.9561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3323 -0.3286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8886 -0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8886 0.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3323 0.9561 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.4447 0.9560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0117 0.6286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5786 0.9560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5786 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0117 1.9380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4447 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8927 0.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1454 1.9379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7803 -0.9707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3047 -0.7281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4621 -0.4281 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.1613 -0.9491 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7398 -0.7838 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3217 -0.9491 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.6226 -0.4281 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0441 -0.5933 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9034 -0.5778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0857 -0.1188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0567 -0.6787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1041 -1.1418 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.7329 -1.6318 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1984 -1.4239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5874 -1.5719 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.0574 -1.0435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5250 -1.2903 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.6615 -1.3447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8556 -2.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8922 -1.9380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3828 -0.7763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7244 -0.1992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1528 0.1602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0400 0.7997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5396 1.2188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4264 1.8607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0169 1.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5381 0.0924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4922 -1.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4922 -2.0956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
35 39 1 0 0 0 0
33 19 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
41 46 2 0 0 0 0
24 47 1 0 0 0 0
47 48 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 47 48
M SBL 2 1 51
M SMT 2 CH2OH
M SVB 2 51 3.6854 -0.9622
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 43 44 45
M SBL 1 1 47
M SMT 1 COOH
M SVB 1 47 -3.9599 1.0045
S SKP 8
ID FL7AAAGL0025
KNApSAcK_ID C00006764
NAME Pelargonidin 3-glucoside-5-(6'''-malonylglucoside)
CAS_RN 167774-88-1
FORMULA C30H33O18
EXACTMASS 681.166689252
AVERAGEMASS 681.57222
SMILES c(c21)c(O[C@@H](C(O)5)O[C@@H]([C@@H](C5O)O)CO)c(c(c4)ccc(O)c4)[o+1]c1cc(cc(O[C@H](O3)[C@@H](O)[C@H]([C@@H](O)C3COC(CC(O)=O)=O)O)2)O
M END