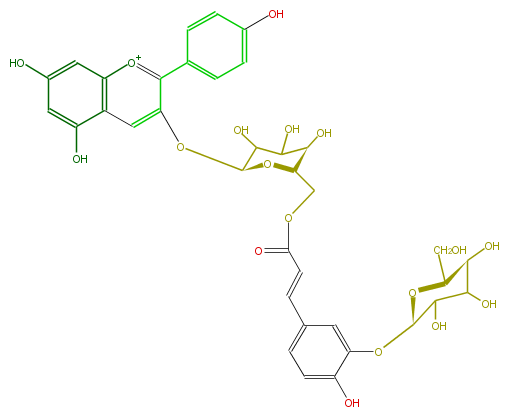
Mol:FL7AAAGL0019
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.4579 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4579 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4579 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4579 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9016 0.2506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9016 0.2506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3453 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3453 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3453 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3453 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9016 1.5353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9016 1.5353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7890 0.2506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7890 0.2506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2327 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2327 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2327 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2327 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7890 1.5353 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -2.7890 1.5353 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6766 1.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6766 1.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1097 1.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1097 1.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5427 1.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5427 1.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5427 2.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5427 2.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1097 2.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1097 2.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6766 2.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6766 2.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0140 1.5352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0140 1.5352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0241 2.5171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0241 2.5171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9016 -0.3916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9016 -0.3916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8166 -0.1489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8166 -0.1489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3138 -0.1727 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.3138 -0.1727 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.5820 -0.6373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5820 -0.6373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0662 -0.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0662 -0.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4527 -0.6373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4527 -0.6373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.7210 -0.1727 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.7210 -0.1727 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2052 -0.3201 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.2052 -0.3201 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.6783 0.1919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6783 0.1919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3469 0.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3469 0.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9827 0.1392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9827 0.1392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8551 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8551 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3385 -1.5667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3385 -1.5667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3386 -2.2306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3386 -2.2306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2531 -2.2307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2531 -2.2307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5854 -2.6505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5854 -2.6505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3292 -3.1556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3292 -3.1556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6607 -3.7297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6607 -3.7297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3640 -4.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3640 -4.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6607 -4.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6607 -4.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2540 -4.7573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2540 -4.7573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5506 -4.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5506 -4.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2539 -3.7297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2539 -3.7297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5503 -5.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5503 -5.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1433 -4.2435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1433 -4.2435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8287 -3.0569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8287 -3.0569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8287 -3.7126 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8287 -3.7126 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2847 -3.2413 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.2847 -3.2413 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.9241 -3.0802 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.9241 -3.0802 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.9242 -2.4244 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.9242 -2.4244 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.4682 -2.8956 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.4682 -2.8956 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 4.3438 -2.1305 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3438 -2.1305 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2785 -3.2848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2785 -3.2848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2847 -3.7235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2847 -3.7235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3191 -2.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3191 -2.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3674 -1.8937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3674 -1.8937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 1 0 0 0 | + | 48 49 1 1 0 0 0 |
| − | 49 44 1 1 0 0 0 | + | 49 44 1 1 0 0 0 |
| − | 48 50 1 0 0 0 0 | + | 48 50 1 0 0 0 0 |
| − | 47 51 1 0 0 0 0 | + | 47 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 49 53 1 0 0 0 0 | + | 49 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 58 3.3956 1.3286 | + | M SVB 1 58 3.3956 1.3286 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0019 | + | ID FL7AAAGL0019 |
| − | KNApSAcK_ID C00006758 | + | KNApSAcK_ID C00006758 |
| − | NAME Pelargonidin 3-[6-(3-glucosylcaffeyl) glucoside] | + | NAME Pelargonidin 3-[6-(3-glucosylcaffeyl) glucoside] |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C36H37O18 | + | FORMULA C36H37O18 |
| − | EXACTMASS 757.1979893800001 | + | EXACTMASS 757.1979893800001 |
| − | AVERAGEMASS 757.66818 | + | AVERAGEMASS 757.66818 |
| − | SMILES c(O)(c6)c(cc(c6)C=CC(=O)OC[C@H]([C@@H]5O)O[C@H](C(C(O)5)O)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)2)c(cc(O)c2)O)O[C@@H](O1)C(C(O)[C@H](O)[C@@H]1CO)O | + | SMILES c(O)(c6)c(cc(c6)C=CC(=O)OC[C@H]([C@@H]5O)O[C@H](C(C(O)5)O)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)2)c(cc(O)c2)O)O[C@@H](O1)C(C(O)[C@H](O)[C@@H]1CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-4.4579 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4579 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9016 0.2506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3453 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3453 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9016 1.5353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7890 0.2506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2327 0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2327 1.2141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7890 1.5353 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-1.6766 1.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1097 1.2078 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5427 1.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5427 2.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1097 2.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6766 2.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0140 1.5352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0241 2.5171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9016 -0.3916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8166 -0.1489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3138 -0.1727 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.5820 -0.6373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0662 -0.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4527 -0.6373 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.7210 -0.1727 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2052 -0.3201 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.6783 0.1919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3469 0.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9827 0.1392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8551 -1.0397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3385 -1.5667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3386 -2.2306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2531 -2.2307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5854 -2.6505 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3292 -3.1556 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6607 -3.7297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3640 -4.2435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6607 -4.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2540 -4.7573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5506 -4.2436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2539 -3.7297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5503 -5.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1433 -4.2435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8287 -3.0569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8287 -3.7126 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2847 -3.2413 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.9241 -3.0802 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.9242 -2.4244 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4682 -2.8956 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
4.3438 -2.1305 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2785 -3.2848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2847 -3.7235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3191 -2.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3674 -1.8937 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
24 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
40 43 1 0 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 1 0 0 0
49 44 1 1 0 0 0
48 50 1 0 0 0 0
47 51 1 0 0 0 0
46 52 1 0 0 0 0
43 45 1 0 0 0 0
49 53 1 0 0 0 0
53 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 58
M SMT 1 CH2OH
M SVB 1 58 3.3956 1.3286
S SKP 8
ID FL7AAAGL0019
KNApSAcK_ID C00006758
NAME Pelargonidin 3-[6-(3-glucosylcaffeyl) glucoside]
CAS_RN -
FORMULA C36H37O18
EXACTMASS 757.1979893800001
AVERAGEMASS 757.66818
SMILES c(O)(c6)c(cc(c6)C=CC(=O)OC[C@H]([C@@H]5O)O[C@H](C(C(O)5)O)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)2)c(cc(O)c2)O)O[C@@H](O1)C(C(O)[C@H](O)[C@@H]1CO)O
M END