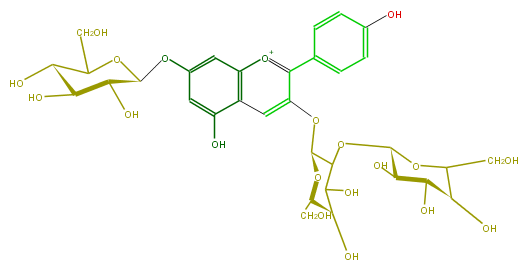
Mol:FL7AAAGL0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2452 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2452 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2452 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2452 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6889 0.2465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6889 0.2465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1326 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1326 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1326 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1326 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6889 1.5312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6889 1.5312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4237 0.2465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4237 0.2465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9800 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9800 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9800 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9800 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4237 1.5312 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.4237 1.5312 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5361 1.5311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5361 1.5311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1031 1.2037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1031 1.2037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6701 1.5311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6701 1.5311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6701 2.1858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6701 2.1858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1031 2.5131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1031 2.5131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5361 2.1858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5361 2.1858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2369 2.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2369 2.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8013 1.5311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8013 1.5311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6889 -0.3957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6889 -0.3957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5565 0.1375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5565 0.1375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4612 -1.6694 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.4612 -1.6694 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.9425 -1.9473 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9425 -1.9473 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7898 -1.4130 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.7898 -1.4130 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.9425 -0.8755 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.9425 -0.8755 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.4612 -0.5976 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.4612 -0.5976 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.6139 -1.1319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6139 -1.1319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2784 -2.8892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2784 -2.8892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2884 -1.4567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2884 -1.4567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1215 -0.3834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1215 -0.3834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2320 -0.4846 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.2320 -0.4846 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 3.3513 -1.1610 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.3513 -1.1610 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.0352 -1.2247 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.0352 -1.2247 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.5978 -1.6186 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.5978 -1.6186 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.4785 -0.9421 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 4.4785 -0.9421 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.7945 -0.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7945 -0.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9359 -0.8701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9359 -0.8701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3672 -2.2574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3672 -2.2574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9796 -1.8608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9796 -1.8608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3183 1.3750 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.3183 1.3750 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.8027 0.6944 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.8027 0.6944 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.0602 0.9831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0602 0.9831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3437 0.9909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.3437 0.9909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8643 1.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8643 1.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5139 1.1688 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.5139 1.1688 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -5.0321 0.9629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0321 0.9629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6065 0.6553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6065 0.6553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6347 0.2689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6347 0.2689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4602 -0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4602 -0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1161 -1.5064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1161 -1.5064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2800 -1.9773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2800 -1.9773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2800 -1.9690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2800 -1.9690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7159 2.1055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7159 2.1055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6679 2.4117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6679 2.4117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 22 27 1 0 0 0 0 | + | 22 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 24 29 1 0 0 0 0 | + | 24 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 30 29 1 0 0 0 0 | + | 30 29 1 0 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 39 1 0 0 0 0 | + | 44 39 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 18 42 1 0 0 0 0 | + | 18 42 1 0 0 0 0 |
| − | 34 48 1 0 0 0 0 | + | 34 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 21 50 1 0 0 0 0 | + | 21 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 44 52 1 0 0 0 0 | + | 44 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 52 53 | + | M SAL 3 2 52 53 |
| − | M SBL 3 1 57 | + | M SBL 3 1 57 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 57 -3.7159 2.1055 | + | M SVB 3 57 -3.7159 2.1055 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 55 | + | M SBL 2 1 55 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 55 1.28 -1.9773 | + | M SVB 2 55 1.28 -1.9773 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 48 49 | + | M SAL 1 2 48 49 |
| − | M SBL 1 1 53 | + | M SBL 1 1 53 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 53 4.8707 0.2092 | + | M SVB 1 53 4.8707 0.2092 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0014 | + | ID FL7AAAGL0014 |
| − | KNApSAcK_ID C00006649 | + | KNApSAcK_ID C00006649 |
| − | NAME Pelargonidin 3-sophoroside-7-glucoside | + | NAME Pelargonidin 3-sophoroside-7-glucoside |
| − | CAS_RN 86279-08-5 | + | CAS_RN 86279-08-5 |
| − | FORMULA C33H41O20 | + | FORMULA C33H41O20 |
| − | EXACTMASS 757.219118752 | + | EXACTMASS 757.219118752 |
| − | AVERAGEMASS 757.6666399999999 | + | AVERAGEMASS 757.6666399999999 |
| − | SMILES Oc(c1)ccc(c([o+1]4)c(O[C@@H](C(O[C@H]([C@@H]6O)OC(CO)[C@@H](O)[C@H]6O)5)O[C@@H]([C@H](O)C5O)CO)cc(c24)c(cc(O[C@H](O3)[C@H]([C@H]([C@H](C3CO)O)O)O)c2)O)c1 | + | SMILES Oc(c1)ccc(c([o+1]4)c(O[C@@H](C(O[C@H]([C@@H]6O)OC(CO)[C@@H](O)[C@H]6O)5)O[C@@H]([C@H](O)C5O)CO)cc(c24)c(cc(O[C@H](O3)[C@H]([C@H]([C@H](C3CO)O)O)O)c2)O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.2452 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2452 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6889 0.2465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1326 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1326 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6889 1.5312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4237 0.2465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9800 0.5676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9800 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4237 1.5312 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.5361 1.5311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1031 1.2037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6701 1.5311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6701 2.1858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1031 2.5131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5361 2.1858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2369 2.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8013 1.5311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6889 -0.3957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5565 0.1375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4612 -1.6694 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.9425 -1.9473 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7898 -1.4130 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9425 -0.8755 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.4612 -0.5976 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6139 -1.1319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2784 -2.8892 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2884 -1.4567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1215 -0.3834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2320 -0.4846 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.3513 -1.1610 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.0352 -1.2247 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.5978 -1.6186 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.4785 -0.9421 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.7945 -0.8785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9359 -0.8701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3672 -2.2574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9796 -1.8608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3183 1.3750 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.8027 0.6944 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.0602 0.9831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3437 0.9909 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8643 1.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5139 1.1688 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-5.0321 0.9629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6065 0.6553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6347 0.2689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4602 -0.7515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1161 -1.5064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2800 -1.9773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2800 -1.9690 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7159 2.1055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6679 2.4117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
25 20 1 0 0 0 0
24 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
31 36 1 0 0 0 0
33 37 1 0 0 0 0
32 38 1 0 0 0 0
30 29 1 0 0 0 0
39 40 1 1 0 0 0
40 41 1 1 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 39 1 0 0 0 0
39 45 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
18 42 1 0 0 0 0
34 48 1 0 0 0 0
48 49 1 0 0 0 0
21 50 1 0 0 0 0
50 51 1 0 0 0 0
44 52 1 0 0 0 0
52 53 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 52 53
M SBL 3 1 57
M SMT 3 CH2OH
M SVB 3 57 -3.7159 2.1055
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 55
M SMT 2 CH2OH
M SVB 2 55 1.28 -1.9773
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 48 49
M SBL 1 1 53
M SMT 1 CH2OH
M SVB 1 53 4.8707 0.2092
S SKP 8
ID FL7AAAGL0014
KNApSAcK_ID C00006649
NAME Pelargonidin 3-sophoroside-7-glucoside
CAS_RN 86279-08-5
FORMULA C33H41O20
EXACTMASS 757.219118752
AVERAGEMASS 757.6666399999999
SMILES Oc(c1)ccc(c([o+1]4)c(O[C@@H](C(O[C@H]([C@@H]6O)OC(CO)[C@@H](O)[C@H]6O)5)O[C@@H]([C@H](O)C5O)CO)cc(c24)c(cc(O[C@H](O3)[C@H]([C@H]([C@H](C3CO)O)O)O)c2)O)c1
M END