Mol:FL6FAFGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 33 36 0 0 0 0 0 0 0 0999 V2000 | + | 33 36 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3359 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3359 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3359 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3359 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8179 -0.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8179 -0.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2999 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2999 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2999 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2999 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8179 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8179 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2181 -0.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2181 -0.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7361 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7361 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7361 0.2381 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.7361 0.2381 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 0.2181 0.5372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2181 0.5372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2541 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2541 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7764 0.2356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7764 0.2356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2987 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2987 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2987 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2987 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7764 1.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7764 1.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2541 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2541 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8539 0.5372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8539 0.5372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0209 -1.2086 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.0209 -1.2086 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.6497 -1.6986 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.6497 -1.6986 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1152 -1.4907 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1152 -1.4907 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5994 -1.4852 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5994 -1.4852 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9742 -1.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9742 -1.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4418 -1.3571 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.4418 -1.3571 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.6118 -1.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6118 -1.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0000 -2.1598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0000 -2.1598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8089 -2.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8089 -2.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8203 -1.2469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8203 -1.2469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0371 2.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0371 2.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4572 2.9124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4572 2.9124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2987 1.1402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2987 1.1402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2987 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2987 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6372 -0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6372 -0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5932 -0.2135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5932 -0.2135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 18 19 1 1 0 0 0 | + | 18 19 1 1 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 18 1 0 0 0 0 | + | 23 18 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 27 3 1 0 0 0 0 | + | 27 3 1 0 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 14 30 1 0 0 0 0 | + | 14 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 23 32 1 0 0 0 0 | + | 23 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 32 33 | + | M SAL 3 2 32 33 |
| − | M SBL 3 1 35 | + | M SBL 3 1 35 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 35 -2.6372 -0.5068 | + | M SVB 3 35 -2.6372 -0.5068 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 30 31 | + | M SAL 2 2 30 31 |
| − | M SBL 2 1 33 | + | M SBL 2 1 33 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 33 2.8202 1.4414 | + | M SVB 2 33 2.8202 1.4414 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 28 29 | + | M SAL 1 2 28 29 |
| − | M SBL 1 1 31 | + | M SBL 1 1 31 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 31 2.0371 2.0049 | + | M SVB 1 31 2.0371 2.0049 |
| − | S SKP 8 | + | S SKP 8 |
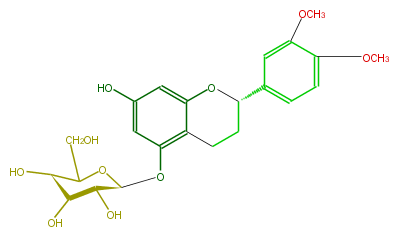
| − | ID FL6FAFGS0001 | + | ID FL6FAFGS0001 |
| − | KNApSAcK_ID C00008768 | + | KNApSAcK_ID C00008768 |
| − | NAME Diffutin | + | NAME Diffutin |
| − | CAS_RN 89289-91-8 | + | CAS_RN 89289-91-8 |
| − | FORMULA C23H28O10 | + | FORMULA C23H28O10 |
| − | EXACTMASS 464.168247116 | + | EXACTMASS 464.168247116 |
| − | AVERAGEMASS 464.46242000000007 | + | AVERAGEMASS 464.46242000000007 |
| − | SMILES c(c4)(c(OC)cc(c4)[C@@H](O3)CCc(c31)c(O[C@@H]([C@@H](O)2)OC(CO)[C@@H]([C@@H]2O)O)cc(O)c1)OC | + | SMILES c(c4)(c(OC)cc(c4)[C@@H](O3)CCc(c31)c(O[C@@H]([C@@H](O)2)OC(CO)[C@@H]([C@@H]2O)O)cc(O)c1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
33 36 0 0 0 0 0 0 0 0999 V2000
-1.3359 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3359 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8179 -0.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2999 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2999 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8179 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2181 -0.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7361 -0.3600 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7361 0.2381 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.2181 0.5372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2541 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7764 0.2356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2987 0.5372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2987 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7764 1.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2541 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8539 0.5372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0209 -1.2086 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.6497 -1.6986 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1152 -1.4907 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5994 -1.4852 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9742 -1.1103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4418 -1.3571 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.6118 -1.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0000 -2.1598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8089 -2.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8203 -1.2469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0371 2.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4572 2.9124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2987 1.1402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2987 1.1402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6372 -0.5068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5932 -0.2135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
18 19 1 1 0 0 0
19 20 1 1 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 18 1 0 0 0 0
18 24 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
27 3 1 0 0 0 0
15 28 1 0 0 0 0
28 29 1 0 0 0 0
14 30 1 0 0 0 0
30 31 1 0 0 0 0
23 32 1 0 0 0 0
32 33 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 32 33
M SBL 3 1 35
M SMT 3 CH2OH
M SVB 3 35 -2.6372 -0.5068
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 30 31
M SBL 2 1 33
M SMT 2 OCH3
M SVB 2 33 2.8202 1.4414
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 28 29
M SBL 1 1 31
M SMT 1 OCH3
M SVB 1 31 2.0371 2.0049
S SKP 8
ID FL6FAFGS0001
KNApSAcK_ID C00008768
NAME Diffutin
CAS_RN 89289-91-8
FORMULA C23H28O10
EXACTMASS 464.168247116
AVERAGEMASS 464.46242000000007
SMILES c(c4)(c(OC)cc(c4)[C@@H](O3)CCc(c31)c(O[C@@H]([C@@H](O)2)OC(CO)[C@@H]([C@@H]2O)O)cc(O)c1)OC
M END