Mol:FL6FA9NC0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 44 0 0 0 0 0 0 0 0999 V2000 | + | 40 44 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.2212 -2.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2212 -2.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7777 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7777 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7777 -1.0454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7777 -1.0454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2212 -0.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2212 -0.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3354 -1.0454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3354 -1.0454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3354 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3354 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3544 -2.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3544 -2.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9310 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9310 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9310 -1.0222 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.9310 -1.0222 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.3544 -0.6893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3544 -0.6893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5477 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5477 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1113 -1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1113 -1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6750 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6750 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6750 -0.1045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6750 -0.1045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1113 0.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1113 0.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5477 -0.1045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5477 -0.1045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7752 -0.7915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7752 -0.7915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2212 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2212 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3417 0.1803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3417 0.1803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3417 0.7793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3417 0.7793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2227 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2227 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2227 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2227 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3417 2.0826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3417 2.0826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9060 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9060 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9060 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9060 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4374 0.7983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4374 0.7983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0245 1.1373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0245 1.1373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0245 1.7534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0245 1.7534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5656 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5656 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1203 1.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1203 1.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6750 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6750 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6750 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6750 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1203 -0.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1203 -0.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5656 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5656 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3417 2.5624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3417 2.5624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7230 2.0456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7230 2.0456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2227 1.1051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2227 1.1051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2227 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2227 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2499 -2.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2499 -2.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4645 -2.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4645 -2.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 5 17 1 0 0 0 0 | + | 5 17 1 0 0 0 0 |
| − | 4 18 1 0 0 0 0 | + | 4 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 2 0 0 0 0 | + | 20 21 2 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 23 35 1 0 0 0 0 | + | 23 35 1 0 0 0 0 |
| − | 22 36 1 0 0 0 0 | + | 22 36 1 0 0 0 0 |
| − | 21 37 1 0 0 0 0 | + | 21 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 1 39 1 0 0 0 0 | + | 1 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 39 40 | + | M SAL 2 2 39 40 |
| − | M SBL 2 1 43 | + | M SBL 2 1 43 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 43 -0.2499 -2.1499 | + | M SVB 2 43 -0.2499 -2.1499 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 41 | + | M SBL 1 1 41 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 41 0.3292 0.8989 | + | M SVB 1 41 0.3292 0.8989 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL6FA9NC0001 | + | ID FL6FA9NC0001 |
| − | KNApSAcK_ID C00008791 | + | KNApSAcK_ID C00008791 |
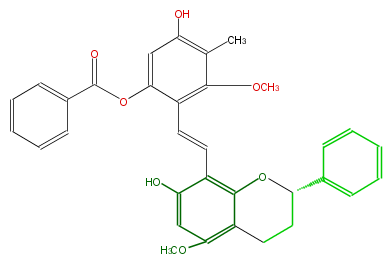
| − | NAME 8-trans-[2-(6-Benzoyloxy-4-hydroxy-2-methoxy-3-methylphenyl)ethenyl]-5-methoxyflavan-7-ol | + | NAME 8-trans-[2-(6-Benzoyloxy-4-hydroxy-2-methoxy-3-methylphenyl)ethenyl]-5-methoxyflavan-7-ol |
| − | CAS_RN 61136-14-9 | + | CAS_RN 61136-14-9 |
| − | FORMULA C33H30O7 | + | FORMULA C33H30O7 |
| − | EXACTMASS 538.199153314 | + | EXACTMASS 538.199153314 |
| − | AVERAGEMASS 538.5871 | + | AVERAGEMASS 538.5871 |
| − | SMILES c(c5)(c(c(OC)c(C)c5O)C=Cc(c32)c(O)cc(OC)c2CC[C@@H](c(c4)cccc4)O3)OC(=O)c(c1)cccc1 | + | SMILES c(c5)(c(c(OC)c(C)c5O)C=Cc(c32)c(O)cc(OC)c2CC[C@@H](c(c4)cccc4)O3)OC(=O)c(c1)cccc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 44 0 0 0 0 0 0 0 0999 V2000
0.2212 -2.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7777 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7777 -1.0454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2212 -0.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3354 -1.0454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3354 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3544 -2.0210 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9310 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9310 -1.0222 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.3544 -0.6893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5477 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1113 -1.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6750 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6750 -0.1045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1113 0.2209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5477 -0.1045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7752 -0.7915 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2212 -0.1447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3417 0.1803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3417 0.7793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2227 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2227 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3417 2.0826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9060 1.7567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9060 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4374 0.7983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0245 1.1373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0245 1.7534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5656 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1203 1.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6750 0.8249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6750 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1203 -0.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5656 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3417 2.5624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7230 2.0456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2227 1.1051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2227 1.1051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2499 -2.1499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4645 -2.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
9 11 1 6 0 0 0
5 17 1 0 0 0 0
4 18 1 0 0 0 0
18 19 2 0 0 0 0
19 20 1 0 0 0 0
20 21 2 0 0 0 0
21 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 20 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
27 29 1 0 0 0 0
29 30 2 0 0 0 0
30 31 1 0 0 0 0
31 32 2 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
34 29 1 0 0 0 0
23 35 1 0 0 0 0
22 36 1 0 0 0 0
21 37 1 0 0 0 0
37 38 1 0 0 0 0
1 39 1 0 0 0 0
39 40 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 39 40
M SBL 2 1 43
M SMT 2 OCH3
M SVB 2 43 -0.2499 -2.1499
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 41
M SMT 1 OCH3
M SVB 1 41 0.3292 0.8989
S SKP 8
ID FL6FA9NC0001
KNApSAcK_ID C00008791
NAME 8-trans-[2-(6-Benzoyloxy-4-hydroxy-2-methoxy-3-methylphenyl)ethenyl]-5-methoxyflavan-7-ol
CAS_RN 61136-14-9
FORMULA C33H30O7
EXACTMASS 538.199153314
AVERAGEMASS 538.5871
SMILES c(c5)(c(c(OC)c(C)c5O)C=Cc(c32)c(O)cc(OC)c2CC[C@@H](c(c4)cccc4)O3)OC(=O)c(c1)cccc1
M END