Mol:FL64AAGM0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5023 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5023 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5023 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5023 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0540 -0.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0540 -0.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6103 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6103 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6103 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6103 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0540 1.0386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0540 1.0386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1666 -0.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1666 -0.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7229 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7229 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7229 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7229 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1666 1.0386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1666 1.0386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2790 1.0385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2790 1.0385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8460 0.7111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8460 0.7111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4130 1.0385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4130 1.0385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4130 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4130 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8460 2.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8460 2.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2790 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2790 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9798 2.0204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9798 2.0204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0540 -0.8882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0540 -0.8882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0584 1.0385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0584 1.0385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1666 -0.8882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1666 -0.8882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0540 1.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0540 1.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4660 0.9619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4660 0.9619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0948 0.4720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0948 0.4720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5603 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5603 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0445 0.6854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0445 0.6854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4193 1.0603 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4193 1.0603 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8869 0.8135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8869 0.8135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9798 0.6653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9798 0.6653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6734 0.4438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6734 0.4438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2540 0.1657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2540 0.1657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0016 -1.2243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0016 -1.2243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6304 -1.7143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6304 -1.7143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0959 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0959 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5802 -1.5008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5802 -1.5008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9549 -1.1260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9549 -1.1260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4226 -1.3728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4226 -1.3728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5154 -1.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5154 -1.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2091 -1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2091 -1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7897 -2.0205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7897 -2.0205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2854 1.5977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2854 1.5977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4885 1.3841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4885 1.3841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2167 0.1665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2167 0.1665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5023 -0.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5023 -0.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8210 -0.5886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8210 -0.5886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0241 -0.8021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0241 -0.8021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 1 0 0 0 | + | 9 11 1 1 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 7 20 1 6 0 0 0 | + | 7 20 1 6 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 27 40 1 0 0 0 0 | + | 27 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 2 42 1 0 0 0 0 | + | 2 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 44 -7.6616 5.7297 | + | M SBV 1 44 -7.6616 5.7297 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 46 -7.9776 5.0370 | + | M SBV 2 46 -7.9776 5.0370 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 ^CH2OH | + | M SMT 3 ^CH2OH |
| − | M SBV 3 48 -7.6616 5.7297 | + | M SBV 3 48 -7.6616 5.7297 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL64AAGM0001 | + | ID FL64AAGM0001 |
| − | KNApSAcK_ID C00008981 | + | KNApSAcK_ID C00008981 |
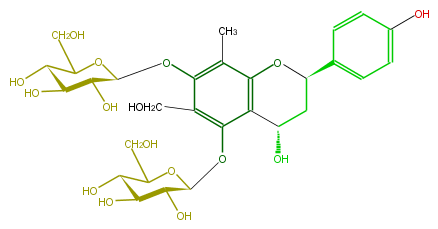
| − | NAME Triphyllin B | + | NAME Triphyllin B |
| − | CAS_RN 101395-03-3 | + | CAS_RN 101395-03-3 |
| − | FORMULA C29H38O16 | + | FORMULA C29H38O16 |
| − | EXACTMASS 642.215985168 | + | EXACTMASS 642.215985168 |
| − | AVERAGEMASS 642.6024199999999 | + | AVERAGEMASS 642.6024199999999 |
| − | SMILES C(C2O)C(Oc(c4C)c2c(c(c(OC(C(O)5)OC(CO)C(C(O)5)O)4)CO)OC(C3O)OC(CO)C(O)C3O)c(c1)ccc(O)c1 | + | SMILES C(C2O)C(Oc(c4C)c2c(c(c(OC(C(O)5)OC(CO)C(C(O)5)O)4)CO)OC(C3O)OC(CO)C(O)C3O)c(c1)ccc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-0.5023 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5023 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0540 -0.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6103 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6103 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0540 1.0386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1666 -0.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7229 0.0751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7229 0.7174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1666 1.0386 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2790 1.0385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8460 0.7111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4130 1.0385 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4130 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8460 2.0205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2790 1.6932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9798 2.0204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0540 -0.8882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0584 1.0385 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1666 -0.8882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0540 1.6807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4660 0.9619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0948 0.4720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5603 0.6798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0445 0.6854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4193 1.0603 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8869 0.8135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9798 0.6653 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6734 0.4438 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2540 0.1657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0016 -1.2243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6304 -1.7143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0959 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5802 -1.5008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9549 -1.1260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4226 -1.3728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5154 -1.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2091 -1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7897 -2.0205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2854 1.5977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4885 1.3841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2167 0.1665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5023 -0.2460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8210 -0.5886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0241 -0.8021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 1 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
7 20 1 6 0 0 0
6 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 18 1 0 0 0 0
25 19 1 0 0 0 0
27 40 1 0 0 0 0
40 41 1 0 0 0 0
2 42 1 0 0 0 0
42 43 1 0 0 0 0
36 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 ^CH2OH
M SBV 1 44 -7.6616 5.7297
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 ^CH2OH
M SBV 2 46 -7.9776 5.0370
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 48
M SMT 3 ^CH2OH
M SBV 3 48 -7.6616 5.7297
S SKP 8
ID FL64AAGM0001
KNApSAcK_ID C00008981
NAME Triphyllin B
CAS_RN 101395-03-3
FORMULA C29H38O16
EXACTMASS 642.215985168
AVERAGEMASS 642.6024199999999
SMILES C(C2O)C(Oc(c4C)c2c(c(c(OC(C(O)5)OC(CO)C(C(O)5)O)4)CO)OC(C3O)OC(CO)C(O)C3O)c(c1)ccc(O)c1
M END