Mol:FL5FGLNS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 30 32 0 0 0 0 0 0 0 0999 V2000 | + | 30 32 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1584 -0.9247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1584 -0.9247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7983 -0.9807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7983 -0.9807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0698 -1.5629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0698 -1.5629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7014 -2.0891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7014 -2.0891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0614 -2.0331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0614 -2.0331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7899 -1.4509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7899 -1.4509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9729 -2.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9729 -2.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6044 -3.1974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6044 -3.1974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9644 -3.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9644 -3.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6930 -2.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6930 -2.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4718 -2.7149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4718 -2.7149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5962 -3.6675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5962 -3.6675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8728 -4.2608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8728 -4.2608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4974 -4.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4974 -4.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1548 -4.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1548 -4.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4316 -4.1467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4316 -4.1467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0560 -3.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0560 -3.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8871 -0.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8871 -0.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7095 -1.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7095 -1.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5303 -5.2762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5303 -5.2762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9200 -5.7033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9200 -5.7033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3427 -6.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3427 -6.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0270 -4.1037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0270 -4.1037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4497 -5.0100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4497 -5.0100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6413 -3.2006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6413 -3.2006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4605 -2.6271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4605 -2.6271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6449 -0.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6449 -0.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1180 -0.9528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1180 -0.9528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1949 -1.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1949 -1.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7395 -2.0343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7395 -2.0343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 8 23 1 0 0 0 0 | + | 8 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 17 25 1 0 0 0 0 | + | 17 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 2 27 1 0 0 0 0 | + | 2 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 6 29 1 0 0 0 0 | + | 6 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 29 30 | + | M SAL 5 2 29 30 |
| − | M SBL 5 1 31 | + | M SBL 5 1 31 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SVB 5 31 -1.2281 0.8998 | + | M SVB 5 31 -1.2281 0.8998 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 27 28 | + | M SAL 4 2 27 28 |
| − | M SBL 4 1 29 | + | M SBL 4 1 29 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 29 -2.7769 -0.5453 | + | M SVB 4 29 -2.7769 -0.5453 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 25 26 | + | M SAL 3 2 25 26 |
| − | M SBL 3 1 27 | + | M SBL 3 1 27 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 27 0.3617 1.6001 | + | M SVB 3 27 0.3617 1.6001 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 23 24 | + | M SAL 2 2 23 24 |
| − | M SBL 2 1 25 | + | M SBL 2 1 25 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 25 0.3617 -0.843 | + | M SVB 2 25 0.3617 -0.843 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 21 22 | + | M SAL 1 2 21 22 |
| − | M SBL 1 1 23 | + | M SBL 1 1 23 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 23 2.0624 0.1204 | + | M SVB 1 23 2.0624 0.1204 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FGLNS0009 | + | ID FL5FGLNS0009 |
| − | KNApSAcK_ID C00004853 | + | KNApSAcK_ID C00004853 |
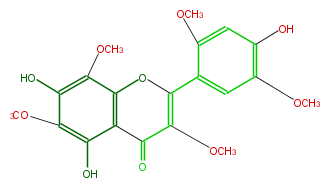
| − | NAME 5,7,4'-Trihydroxy-3,6,8,2',5'-pentamethoxyflavone | + | NAME 5,7,4'-Trihydroxy-3,6,8,2',5'-pentamethoxyflavone |
| − | CAS_RN 107644-09-7 | + | CAS_RN 107644-09-7 |
| − | FORMULA C20H20O10 | + | FORMULA C20H20O10 |
| − | EXACTMASS 420.10564686 | + | EXACTMASS 420.10564686 |
| − | AVERAGEMASS 420.3668 | + | AVERAGEMASS 420.3668 |
| − | SMILES COc(c1)c(C(O3)=C(C(c(c32)c(c(c(O)c2OC)OC)O)=O)OC)cc(c(O)1)OC | + | SMILES COc(c1)c(C(O3)=C(C(c(c32)c(c(c(O)c2OC)OC)O)=O)OC)cc(c(O)1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
30 32 0 0 0 0 0 0 0 0999 V2000
-1.1584 -0.9247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7983 -0.9807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0698 -1.5629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7014 -2.0891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0614 -2.0331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7899 -1.4509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9729 -2.6713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6044 -3.1974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9644 -3.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6930 -2.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4718 -2.7149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5962 -3.6675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8728 -4.2608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4974 -4.7970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1548 -4.7400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4316 -4.1467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0560 -3.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8871 -0.3428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7095 -1.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5303 -5.2762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9200 -5.7033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3427 -6.6096 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0270 -4.1037 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4497 -5.0100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6413 -3.2006 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4605 -2.6271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6449 -0.2769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1180 -0.9528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1949 -1.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7395 -2.0343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
14 21 1 0 0 0 0
21 22 1 0 0 0 0
8 23 1 0 0 0 0
23 24 1 0 0 0 0
17 25 1 0 0 0 0
25 26 1 0 0 0 0
2 27 1 0 0 0 0
27 28 1 0 0 0 0
6 29 1 0 0 0 0
29 30 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 29 30
M SBL 5 1 31
M SMT 5 OCH3
M SVB 5 31 -1.2281 0.8998
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 27 28
M SBL 4 1 29
M SMT 4 OCH3
M SVB 4 29 -2.7769 -0.5453
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 25 26
M SBL 3 1 27
M SMT 3 OCH3
M SVB 3 27 0.3617 1.6001
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 23 24
M SBL 2 1 25
M SMT 2 OCH3
M SVB 2 25 0.3617 -0.843
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 21 22
M SBL 1 1 23
M SMT 1 OCH3
M SVB 1 23 2.0624 0.1204
S SKP 8
ID FL5FGLNS0009
KNApSAcK_ID C00004853
NAME 5,7,4'-Trihydroxy-3,6,8,2',5'-pentamethoxyflavone
CAS_RN 107644-09-7
FORMULA C20H20O10
EXACTMASS 420.10564686
AVERAGEMASS 420.3668
SMILES COc(c1)c(C(O3)=C(C(c(c32)c(c(c(O)c2OC)OC)O)=O)OC)cc(c(O)1)OC
M END