Mol:FL5FGGNS0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 30 32 0 0 0 0 0 0 0 0999 V2000 | + | 30 32 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6513 -0.2423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6513 -0.2423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3798 -0.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3798 -0.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7399 -0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7399 -0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3715 -0.3542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3715 -0.3542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6429 0.2279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6429 0.2279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2828 0.2839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2828 0.2839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7315 -0.4102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7315 -0.4102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3631 0.1160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3631 0.1160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6346 0.6981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6346 0.6981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2745 0.7541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2745 0.7541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5199 -0.8641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5199 -0.8641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2663 1.2242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2663 1.2242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3859 1.1671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3859 1.1671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7614 1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7614 1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4847 2.2967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4847 2.2967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1675 2.3537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1675 2.3537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5429 1.8175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5429 1.8175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2766 0.0600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2766 0.0600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4685 -1.4623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4685 -1.4623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4441 2.9469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4441 2.9469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0936 -0.9210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0936 -0.9210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2966 -1.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2966 -1.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2730 0.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2730 0.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2577 1.9206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2577 1.9206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2366 0.1675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2366 0.1675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0558 0.7411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0558 0.7411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8601 2.8328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8601 2.8328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6820 2.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6820 2.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7576 1.6162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7576 1.6162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7538 1.5290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7538 1.5290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 2 21 1 0 0 0 0 | + | 2 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 1 25 1 0 0 0 0 | + | 1 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 15 27 1 0 0 0 0 | + | 15 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 14 29 1 0 0 0 0 | + | 14 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 29 30 | + | M SAL 5 2 29 30 |
| − | M SBL 5 1 31 | + | M SBL 5 1 31 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SVB 5 31 1.8838 -0.0612 | + | M SVB 5 31 1.8838 -0.0612 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 27 28 | + | M SAL 4 2 27 28 |
| − | M SBL 4 1 29 | + | M SBL 4 1 29 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 29 2.241 1.127 | + | M SVB 4 29 2.241 1.127 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 25 26 | + | M SAL 3 2 25 26 |
| − | M SBL 3 1 27 | + | M SBL 3 1 27 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 27 -2.5983 0.4428 | + | M SVB 3 27 -2.5983 0.4428 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 23 24 | + | M SAL 2 2 23 24 |
| − | M SBL 2 1 25 | + | M SBL 2 1 25 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 25 -1.4067 0.7182 | + | M SVB 2 25 -1.4067 0.7182 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 21 22 | + | M SAL 1 2 21 22 |
| − | M SBL 1 1 23 | + | M SBL 1 1 23 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 23 -2.9555 -0.7269 | + | M SVB 1 23 -2.9555 -0.7269 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FGGNS0008 | + | ID FL5FGGNS0008 |
| − | KNApSAcK_ID C00004865 | + | KNApSAcK_ID C00004865 |
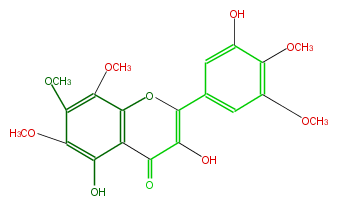
| − | NAME 3-Demethyldigicitrin;3,5,3'-Trihydroxy-6,7,8,4',5'-pentamethoxyflavone;3,5-Dihydroxy-2-(3-hydroxy-4,5-dimethoxyphenyl)-6,7,8-trimethoxy-4H-1-benzopyran-4-one | + | NAME 3-Demethyldigicitrin;3,5,3'-Trihydroxy-6,7,8,4',5'-pentamethoxyflavone;3,5-Dihydroxy-2-(3-hydroxy-4,5-dimethoxyphenyl)-6,7,8-trimethoxy-4H-1-benzopyran-4-one |
| − | CAS_RN 158437-61-7 | + | CAS_RN 158437-61-7 |
| − | FORMULA C20H20O10 | + | FORMULA C20H20O10 |
| − | EXACTMASS 420.10564686 | + | EXACTMASS 420.10564686 |
| − | AVERAGEMASS 420.3668 | + | AVERAGEMASS 420.3668 |
| − | SMILES COc(c(O)3)c(cc(c3)C(O2)=C(C(c(c21)c(c(c(c(OC)1)OC)OC)O)=O)O)OC | + | SMILES COc(c(O)3)c(cc(c3)C(O2)=C(C(c(c21)c(c(c(c(OC)1)OC)OC)O)=O)O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
30 32 0 0 0 0 0 0 0 0999 V2000
-2.6513 -0.2423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3798 -0.8244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7399 -0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3715 -0.3542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6429 0.2279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2828 0.2839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7315 -0.4102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3631 0.1160 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6346 0.6981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2745 0.7541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5199 -0.8641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2663 1.2242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3859 1.1671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7614 1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4847 2.2967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1675 2.3537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5429 1.8175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2766 0.0600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4685 -1.4623 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4441 2.9469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0936 -0.9210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2966 -1.1344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2730 0.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2577 1.9206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2366 0.1675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0558 0.7411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8601 2.8328 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6820 2.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7576 1.6162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7538 1.5290 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
8 18 1 0 0 0 0
3 19 1 0 0 0 0
16 20 1 0 0 0 0
2 21 1 0 0 0 0
21 22 1 0 0 0 0
6 23 1 0 0 0 0
23 24 1 0 0 0 0
1 25 1 0 0 0 0
25 26 1 0 0 0 0
15 27 1 0 0 0 0
27 28 1 0 0 0 0
14 29 1 0 0 0 0
29 30 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 29 30
M SBL 5 1 31
M SMT 5 OCH3
M SVB 5 31 1.8838 -0.0612
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 27 28
M SBL 4 1 29
M SMT 4 OCH3
M SVB 4 29 2.241 1.127
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 25 26
M SBL 3 1 27
M SMT 3 OCH3
M SVB 3 27 -2.5983 0.4428
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 23 24
M SBL 2 1 25
M SMT 2 OCH3
M SVB 2 25 -1.4067 0.7182
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 21 22
M SBL 1 1 23
M SMT 1 OCH3
M SVB 1 23 -2.9555 -0.7269
S SKP 8
ID FL5FGGNS0008
KNApSAcK_ID C00004865
NAME 3-Demethyldigicitrin;3,5,3'-Trihydroxy-6,7,8,4',5'-pentamethoxyflavone;3,5-Dihydroxy-2-(3-hydroxy-4,5-dimethoxyphenyl)-6,7,8-trimethoxy-4H-1-benzopyran-4-one
CAS_RN 158437-61-7
FORMULA C20H20O10
EXACTMASS 420.10564686
AVERAGEMASS 420.3668
SMILES COc(c(O)3)c(cc(c3)C(O2)=C(C(c(c21)c(c(c(c(OC)1)OC)OC)O)=O)O)OC
M END