Mol:FL5FFCGL0011
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 51 0 0 0 0 0 0 0 0999 V2000 | + | 47 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6903 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6903 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6903 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6903 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1340 -0.0543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1340 -0.0543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5777 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5777 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5777 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5777 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1340 1.2304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1340 1.2304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0214 -0.0543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0214 -0.0543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4651 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4651 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4651 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4651 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0214 1.2304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0214 1.2304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0214 -0.5552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0214 -0.5552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2354 1.3540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2354 1.3540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8024 1.0267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8024 1.0267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3694 1.3540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3694 1.3540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3694 2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3694 2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8024 2.3361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8024 2.3361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2354 2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2354 2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1340 -0.6965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1340 -0.6965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2354 2.5087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2354 2.5087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3548 1.2929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3548 1.2929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0211 -0.1771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0211 -0.1771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5995 -1.2906 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.5995 -1.2906 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.0463 -0.9712 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.0463 -0.9712 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2218 -1.5855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2218 -1.5855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0463 -2.2034 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.0463 -2.2034 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.5995 -2.5229 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.5995 -2.5229 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4240 -1.9086 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.4240 -1.9086 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.4406 -0.6974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4406 -0.6974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9279 -1.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9279 -1.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9353 -3.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9353 -3.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1739 -0.2191 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.1739 -0.2191 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.5756 -0.7494 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.5756 -0.7494 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1541 -0.5244 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1541 -0.5244 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7122 -0.5184 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7122 -0.5184 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3066 -0.1127 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3066 -0.1127 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.8006 -0.3798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8006 -0.3798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9494 -0.7799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9494 -0.7799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4855 -1.0808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4855 -1.0808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8982 -0.7840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8982 -0.7840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8559 1.8034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8559 1.8034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4193 2.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4193 2.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3031 -2.2993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3031 -2.2993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5563 -3.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5563 -3.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0858 2.9122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0858 2.9122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5272 3.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5272 3.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4133 0.2042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4133 0.2042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9967 0.7876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9967 0.7876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 6 40 1 0 0 0 0 | + | 6 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 35 46 1 0 0 0 0 | + | 35 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 46 47 | + | M SAL 4 2 46 47 |
| − | M SBL 4 1 50 | + | M SBL 4 1 50 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 50 2.6403 -0.1338 | + | M SVB 4 50 2.6403 -0.1338 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 46 | + | M SBL 3 1 46 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 46 -0.185 -2.4824 | + | M SVB 3 46 -0.185 -2.4824 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 48 1.0858 2.9122 | + | M SVB 2 48 1.0858 2.9122 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 44 -1.8559 1.8034 | + | M SVB 1 44 -1.8559 1.8034 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FFCGL0011 | + | ID FL5FFCGL0011 |
| − | KNApSAcK_ID C00005718 | + | KNApSAcK_ID C00005718 |
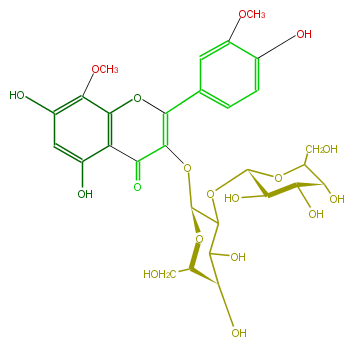
| − | NAME Limocitrin 3-sophoroside | + | NAME Limocitrin 3-sophoroside |
| − | CAS_RN 114882-19-8 | + | CAS_RN 114882-19-8 |
| − | FORMULA C29H34O18 | + | FORMULA C29H34O18 |
| − | EXACTMASS 670.174514284 | + | EXACTMASS 670.174514284 |
| − | AVERAGEMASS 670.5694599999999 | + | AVERAGEMASS 670.5694599999999 |
| − | SMILES O[C@H](C5CO)[C@@H](O)[C@@H](O)[C@H](O5)OC([C@@H]1OC(C3=O)=C(c(c4)ccc(O)c(OC)4)Oc(c23)c(c(O)cc(O)2)OC)C([C@@H](O)[C@@H](CO)O1)O | + | SMILES O[C@H](C5CO)[C@@H](O)[C@@H](O)[C@H](O5)OC([C@@H]1OC(C3=O)=C(c(c4)ccc(O)c(OC)4)Oc(c23)c(c(O)cc(O)2)OC)C([C@@H](O)[C@@H](CO)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 51 0 0 0 0 0 0 0 0999 V2000
-2.6903 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6903 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1340 -0.0543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5777 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5777 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1340 1.2304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0214 -0.0543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4651 0.2669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4651 0.9092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0214 1.2304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0214 -0.5552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2354 1.3540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8024 1.0267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3694 1.3540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3694 2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8024 2.3361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2354 2.0087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1340 -0.6965 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2354 2.5087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3548 1.2929 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0211 -0.1771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5995 -1.2906 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.0463 -0.9712 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2218 -1.5855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0463 -2.2034 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.5995 -2.5229 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4240 -1.9086 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4406 -0.6974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9279 -1.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9353 -3.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1739 -0.2191 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.5756 -0.7494 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1541 -0.5244 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7122 -0.5184 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3066 -0.1127 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.8006 -0.3798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9494 -0.7799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4855 -1.0808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8982 -0.7840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8559 1.8034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4193 2.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3031 -2.2993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5563 -3.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0858 2.9122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5272 3.8095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4133 0.2042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9967 0.7876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
28 31 1 0 0 0 0
6 40 1 0 0 0 0
40 41 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
16 44 1 0 0 0 0
44 45 1 0 0 0 0
35 46 1 0 0 0 0
46 47 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 46 47
M SBL 4 1 50
M SMT 4 CH2OH
M SVB 4 50 2.6403 -0.1338
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 46
M SMT 3 CH2OH
M SVB 3 46 -0.185 -2.4824
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 48
M SMT 2 OCH3
M SVB 2 48 1.0858 2.9122
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 OCH3
M SVB 1 44 -1.8559 1.8034
S SKP 8
ID FL5FFCGL0011
KNApSAcK_ID C00005718
NAME Limocitrin 3-sophoroside
CAS_RN 114882-19-8
FORMULA C29H34O18
EXACTMASS 670.174514284
AVERAGEMASS 670.5694599999999
SMILES O[C@H](C5CO)[C@@H](O)[C@@H](O)[C@H](O5)OC([C@@H]1OC(C3=O)=C(c(c4)ccc(O)c(OC)4)Oc(c23)c(c(O)cc(O)2)OC)C([C@@H](O)[C@@H](CO)O1)O
M END