Mol:FL5FFAGS0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.9991 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9991 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9990 0.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9990 0.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2846 -0.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2846 -0.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5701 0.2651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5701 0.2651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5702 1.0901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5702 1.0901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2846 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2846 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1443 -0.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1443 -0.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8586 0.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8586 0.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8587 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8587 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1443 1.5027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1443 1.5027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1443 -0.9446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1443 -0.9446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5934 1.5583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5934 1.5583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3214 1.1379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3214 1.1379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0496 1.5582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0496 1.5582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0497 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0497 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3215 2.8194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3215 2.8194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5933 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5933 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2846 -0.9624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2846 -0.9624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8290 2.8489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8290 2.8489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6612 -0.1989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6612 -0.1989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7203 1.5066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7203 1.5066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2832 2.3259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2832 2.3259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1379 4.2567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1379 4.2567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2705 3.4356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2705 3.4356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5816 3.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5816 3.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0784 2.5350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0784 2.5350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9195 3.3562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9195 3.3562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7141 3.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7141 3.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8290 4.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8290 4.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7709 4.0325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7709 4.0325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5816 2.2761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5816 2.2761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2675 -1.2066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2675 -1.2066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5020 -0.8842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5020 -0.8842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9186 -1.6693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9186 -1.6693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9070 -2.4461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9070 -2.4461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6533 -2.6462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6533 -2.6462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2559 -1.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2559 -1.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1554 -0.4727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1554 -0.4727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0042 -2.3117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0042 -2.3117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5965 -3.2088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5965 -3.2088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0665 -3.6201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0665 -3.6201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9552 -4.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9552 -4.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8193 -3.9259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8193 -3.9259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6508 4.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6508 4.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7332 3.7515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7332 3.7515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 6 22 1 0 0 0 0 | + | 6 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 22 26 1 0 0 0 0 | + | 22 26 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 20 33 1 0 0 0 0 | + | 20 33 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 28 44 1 0 0 0 0 | + | 28 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 41 42 43 | + | M SAL 1 3 41 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 47 -0.1596 1.1741 | + | M SBV 1 47 -0.1596 1.1741 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 49 -0.0633 -0.7661 | + | M SBV 2 49 -0.0633 -0.7661 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FFAGS0010 | + | ID FL5FFAGS0010 |
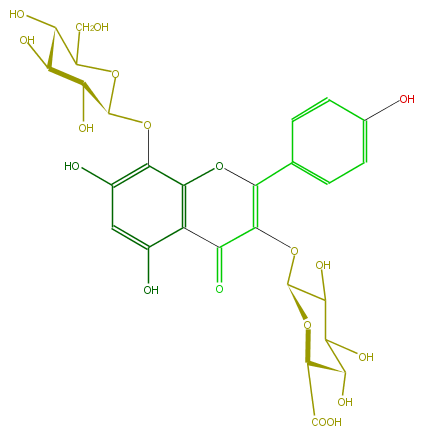
| − | FORMULA C27H28O18 | + | FORMULA C27H28O18 |
| − | EXACTMASS 640.1275640920001 | + | EXACTMASS 640.1275640920001 |
| − | AVERAGEMASS 640.5004200000001 | + | AVERAGEMASS 640.5004200000001 |
| − | SMILES C(Oc(c42)c(cc(O)c2C(=O)C(OC(C5O)OC(C(C5O)O)C(O)=O)=C(O4)c(c3)ccc(O)c3)O)(C1O)OC(C(C(O)1)O)CO | + | SMILES C(Oc(c42)c(cc(O)c2C(=O)C(OC(C5O)OC(C(C5O)O)C(O)=O)=C(O4)c(c3)ccc(O)c3)O)(C1O)OC(C(C(O)1)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-1.9991 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9990 0.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2846 -0.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5701 0.2651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5702 1.0901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2846 1.5027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1443 -0.1473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8586 0.2652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8587 1.0902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1443 1.5027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1443 -0.9446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5934 1.5583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3214 1.1379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0496 1.5582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0497 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3215 2.8194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5933 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2846 -0.9624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8290 2.8489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6612 -0.1989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7203 1.5066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2832 2.3259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1379 4.2567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2705 3.4356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5816 3.1443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0784 2.5350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9195 3.3562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7141 3.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8290 4.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7709 4.0325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5816 2.2761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2675 -1.2066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5020 -0.8842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9186 -1.6693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9070 -2.4461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6533 -2.6462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2559 -1.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1554 -0.4727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0042 -2.3117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5965 -3.2088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0665 -3.6201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9552 -4.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8193 -3.9259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6508 4.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7332 3.7515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
1 21 1 0 0 0 0
6 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
22 26 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
20 33 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
35 41 1 0 0 0 0
44 45 1 0 0 0 0
28 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 41 42 43
M SBL 1 1 47
M SMT 1 COOH
M SBV 1 47 -0.1596 1.1741
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 49
M SMT 2 CH2OH
M SBV 2 49 -0.0633 -0.7661
S SKP 5
ID FL5FFAGS0010
FORMULA C27H28O18
EXACTMASS 640.1275640920001
AVERAGEMASS 640.5004200000001
SMILES C(Oc(c42)c(cc(O)c2C(=O)C(OC(C5O)OC(C(C5O)O)C(O)=O)=C(O4)c(c3)ccc(O)c3)O)(C1O)OC(C(C(O)1)O)CO
M END