Mol:FL5FECGS0068
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | 2.0910 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0910 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3766 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3766 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3766 2.3807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3766 2.3807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0910 2.7932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0910 2.7932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8055 2.3807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8055 2.3807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8055 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8055 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6621 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6621 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0524 1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0524 1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7669 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7669 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7669 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7669 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0524 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0524 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6621 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6621 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4813 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4813 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1958 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1958 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1958 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1958 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4813 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4813 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0524 -0.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0524 -0.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9103 1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9103 1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3766 -0.0943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3766 -0.0943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4813 -0.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4813 -0.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5200 2.7932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5200 2.7932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0910 3.6182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0910 3.6182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8569 -0.0635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8569 -0.0635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8349 4.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8349 4.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5200 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5200 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5035 1.2132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5035 1.2132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1269 -2.1388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1269 -2.1388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6793 -2.3151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6793 -2.3151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0624 -1.5841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0624 -1.5841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7645 -1.1504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7645 -1.1504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9583 -0.9740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9583 -0.9740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5751 -1.7050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5751 -1.7050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0026 -1.4063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0026 -1.4063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5308 -1.5745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5308 -1.5745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6180 -2.1616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6180 -2.1616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0749 -2.7986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0749 -2.7986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6449 -1.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6449 -1.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5589 -2.9021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5589 -2.9021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1462 -3.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1462 -3.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3479 -3.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3479 -3.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5544 -3.6346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5544 -3.6346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9670 -2.9198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9670 -2.9198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7654 -3.1289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7654 -3.1289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9223 -2.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9223 -2.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4022 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4022 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0421 -3.3853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0421 -3.3853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5949 -3.3578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5949 -3.3578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5194 -4.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5194 -4.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 15 23 1 0 0 0 0 | + | 15 23 1 0 0 0 0 |
| − | 22 24 1 0 0 0 0 | + | 22 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 18 26 1 0 0 0 0 | + | 18 26 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 27 35 1 0 0 0 0 | + | 27 35 1 0 0 0 0 |
| − | 28 36 1 0 0 0 0 | + | 28 36 1 0 0 0 0 |
| − | 29 37 1 0 0 0 0 | + | 29 37 1 0 0 0 0 |
| − | 30 19 1 0 0 0 0 | + | 30 19 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 38 1 0 0 0 0 | + | 43 38 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 38 46 1 0 0 0 0 | + | 38 46 1 0 0 0 0 |
| − | 39 47 1 0 0 0 0 | + | 39 47 1 0 0 0 0 |
| − | 40 48 1 0 0 0 0 | + | 40 48 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGS0068 | + | ID FL5FECGS0068 |
| − | KNApSAcK_ID C00013958 | + | KNApSAcK_ID C00013958 |
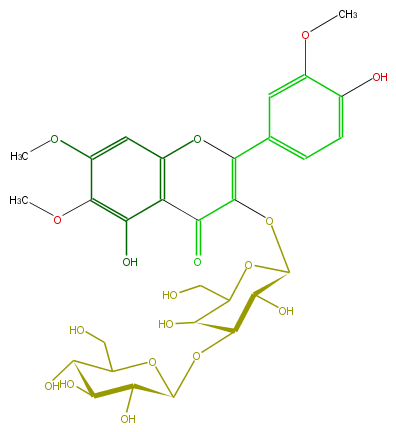
| − | NAME Veronicafolin 3-glucosyl-(1->3)-galactoside;Quercetagetin 6,7,3'-trimethyl ether 3-glucosyl-(1->3)-galactoside | + | NAME Veronicafolin 3-glucosyl-(1->3)-galactoside;Quercetagetin 6,7,3'-trimethyl ether 3-glucosyl-(1->3)-galactoside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C30H36O18 | + | FORMULA C30H36O18 |
| − | EXACTMASS 684.190164348 | + | EXACTMASS 684.190164348 |
| − | AVERAGEMASS 684.59604 | + | AVERAGEMASS 684.59604 |
| − | SMILES COc(c1O)c(OC)cc(O4)c(C(C(=C4c(c5)cc(OC)c(c5)O)OC(O2)C(O)C(OC(C(O)3)OC(C(O)C3O)CO)C(O)C2CO)=O)1 | + | SMILES COc(c1O)c(OC)cc(O4)c(C(C(=C4c(c5)cc(OC)c(c5)O)OC(O2)C(O)C(OC(C(O)3)OC(C(O)C3O)CO)C(O)C2CO)=O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
2.0910 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3766 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3766 2.3807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0910 2.7932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8055 2.3807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8055 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6621 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0524 1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7669 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7669 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0524 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6621 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4813 1.5557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1958 1.1432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1958 0.3182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4813 -0.0943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0524 -0.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9103 1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3766 -0.0943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4813 -0.9193 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5200 2.7932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0910 3.6182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8569 -0.0635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8349 4.0476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5200 0.3193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5035 1.2132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1269 -2.1388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6793 -2.3151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0624 -1.5841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7645 -1.1504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9583 -0.9740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5751 -1.7050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0026 -1.4063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5308 -1.5745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6180 -2.1616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0749 -2.7986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6449 -1.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5589 -2.9021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1462 -3.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3479 -3.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5544 -3.6346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9670 -2.9198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7654 -3.1289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9223 -2.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4022 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0421 -3.3853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5949 -3.3578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5194 -4.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
15 23 1 0 0 0 0
22 24 1 0 0 0 0
23 25 1 0 0 0 0
18 26 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
27 35 1 0 0 0 0
28 36 1 0 0 0 0
29 37 1 0 0 0 0
30 19 1 0 0 0 0
38 39 1 1 0 0 0
39 40 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 38 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
38 46 1 0 0 0 0
39 47 1 0 0 0 0
40 48 1 0 0 0 0
41 36 1 0 0 0 0
S SKP 8
ID FL5FECGS0068
KNApSAcK_ID C00013958
NAME Veronicafolin 3-glucosyl-(1->3)-galactoside;Quercetagetin 6,7,3'-trimethyl ether 3-glucosyl-(1->3)-galactoside
CAS_RN -
FORMULA C30H36O18
EXACTMASS 684.190164348
AVERAGEMASS 684.59604
SMILES COc(c1O)c(OC)cc(O4)c(C(C(=C4c(c5)cc(OC)c(c5)O)OC(O2)C(O)C(OC(C(O)3)OC(C(O)C3O)CO)C(O)C2CO)=O)1
M END