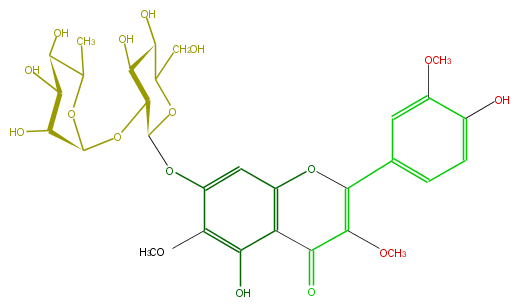
Mol:FL5FECGS0028
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 51 0 0 0 0 0 0 0 0999 V2000 | + | 47 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1144 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1144 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1144 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1144 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3998 -1.9089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3998 -1.9089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3147 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3147 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3147 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3147 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3998 -0.2588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3998 -0.2588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0292 -1.9089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0292 -1.9089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7438 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7438 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7438 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7438 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0292 -0.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0292 -0.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0292 -2.7100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0292 -2.7100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6434 -0.0999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6434 -0.0999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3717 -0.5205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3717 -0.5205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1000 -0.0999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1000 -0.0999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1000 0.7409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1000 0.7409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3717 1.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3717 1.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6434 0.7409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6434 0.7409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3998 -2.7337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3998 -2.7337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7819 1.1346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7819 1.1346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7929 -0.2947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7929 -0.2947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2134 0.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2134 0.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5388 0.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5388 0.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7529 0.8485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7529 0.8485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5388 1.6021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5388 1.6021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2134 1.9917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2134 1.9917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9994 1.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9994 1.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0419 2.4761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0419 2.4761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5887 2.3083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5887 2.3083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6254 1.7339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6254 1.7339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7819 0.4793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7819 0.4793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0912 2.2193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0912 2.2193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5935 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5935 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2418 1.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2418 1.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2418 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2418 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7394 0.8920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7394 0.8920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0912 1.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0912 1.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8331 0.3796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8331 0.3796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7435 2.3455 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7435 2.3455 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3112 2.8515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3112 2.8515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4880 -1.9260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4880 -1.9260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0371 -3.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0371 -3.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3717 1.9346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3717 1.9346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9386 3.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9386 3.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6818 2.1404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6818 2.1404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8700 1.4593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8700 1.4593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8461 -1.9188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8461 -1.9188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6578 -2.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6578 -2.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 21 30 1 0 0 0 0 | + | 21 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 37 22 1 0 0 0 0 | + | 37 22 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 31 39 1 0 0 0 0 | + | 31 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 8 40 1 0 0 0 0 | + | 8 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 2 46 1 0 0 0 0 | + | 2 46 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 45 -0.7442 0.4297 | + | M SBV 1 45 -0.7442 0.4297 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 0.0000 -0.7732 | + | M SBV 2 47 0.0000 -0.7732 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 49 | + | M SBL 3 1 49 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 49 -0.4095 -0.6312 | + | M SBV 3 49 -0.4095 -0.6312 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 46 47 | + | M SAL 4 2 46 47 |
| − | M SBL 4 1 51 | + | M SBL 4 1 51 |
| − | M SMT 4 ^ OCH3 | + | M SMT 4 ^ OCH3 |
| − | M SBV 4 51 0.7317 0.4225 | + | M SBV 4 51 0.7317 0.4225 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FECGS0028 | + | ID FL5FECGS0028 |
| − | FORMULA C30H36O17 | + | FORMULA C30H36O17 |
| − | EXACTMASS 668.1952497259999 | + | EXACTMASS 668.1952497259999 |
| − | AVERAGEMASS 668.59664 | + | AVERAGEMASS 668.59664 |
| − | SMILES c(c3OC(O4)C(OC(O5)C(O)C(C(O)C5C)O)C(C(C4CO)O)O)(c(c(c(c3)1)C(=O)C(=C(c(c2)ccc(c2OC)O)O1)OC)O)OC | + | SMILES c(c3OC(O4)C(OC(O5)C(O)C(C(O)C5C)O)C(C(C4CO)O)O)(c(c(c(c3)1)C(=O)C(=C(c(c2)ccc(c2OC)O)O1)OC)O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 51 0 0 0 0 0 0 0 0999 V2000
-1.1144 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1144 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3998 -1.9089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3147 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3147 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3998 -0.2588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0292 -1.9089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7438 -1.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7438 -0.6714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0292 -0.2588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0292 -2.7100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6434 -0.0999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3717 -0.5205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1000 -0.0999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1000 0.7409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3717 1.1614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6434 0.7409 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3998 -2.7337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7819 1.1346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7929 -0.2947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2134 0.4890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5388 0.0996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7529 0.8485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5388 1.6021 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2134 1.9917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9994 1.2426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0419 2.4761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5887 2.3083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6254 1.7339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7819 0.4793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0912 2.2193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5935 1.7170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2418 1.1000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2418 0.3900 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7394 0.8920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0912 1.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8331 0.3796 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7435 2.3455 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3112 2.8515 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4880 -1.9260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0371 -3.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3717 1.9346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9386 3.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6818 2.1404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8700 1.4593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8461 -1.9188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6578 -2.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
26 29 1 0 0 0 0
21 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
34 20 1 0 0 0 0
33 37 1 0 0 0 0
37 22 1 0 0 0 0
32 38 1 0 0 0 0
31 39 1 0 0 0 0
40 41 1 0 0 0 0
8 40 1 0 0 0 0
42 43 1 0 0 0 0
16 42 1 0 0 0 0
44 45 1 0 0 0 0
36 44 1 0 0 0 0
46 47 1 0 0 0 0
2 46 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 OCH3
M SBV 1 45 -0.7442 0.4297
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 0.0000 -0.7732
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 49
M SMT 3 CH2OH
M SBV 3 49 -0.4095 -0.6312
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 46 47
M SBL 4 1 51
M SMT 4 ^ OCH3
M SBV 4 51 0.7317 0.4225
S SKP 5
ID FL5FECGS0028
FORMULA C30H36O17
EXACTMASS 668.1952497259999
AVERAGEMASS 668.59664
SMILES c(c3OC(O4)C(OC(O5)C(O)C(C(O)C5C)O)C(C(C4CO)O)O)(c(c(c(c3)1)C(=O)C(=C(c(c2)ccc(c2OC)O)O1)OC)O)OC
M END