Mol:FL5FEAGS0025
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 40 0 0 0 0 0 0 0 0999 V2000 | + | 36 40 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0784 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0784 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6361 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6361 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6361 1.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6361 1.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0784 1.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0784 1.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7929 1.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7929 1.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7929 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7929 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3504 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3504 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0649 0.5071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0649 0.5071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7794 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7794 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7794 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7794 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0649 -1.1429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0649 -1.1429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3504 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3504 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4939 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4939 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2083 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2083 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2083 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2083 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4939 -1.1429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4939 -1.1429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0649 -1.8499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0649 -1.8499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4939 -1.8834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4939 -1.8834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9928 0.3495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9928 0.3495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4778 -0.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4778 -0.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9928 -0.9854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9928 -0.9854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6591 1.7635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6591 1.7635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0538 1.2026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0538 1.2026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8789 1.1849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8789 1.1849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5797 0.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5797 0.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1852 1.3100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1852 1.3100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3600 1.3278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3600 1.3278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0903 1.7725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0903 1.7725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9803 0.9992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9803 0.9992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0228 1.8834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0228 1.8834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3963 1.6804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3963 1.6804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5006 0.6259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5006 0.6259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4778 0.8376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4778 0.8376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1952 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1952 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6799 -1.1176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6799 -1.1176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6799 -1.8520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6799 -1.8520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 5 31 1 0 0 0 0 | + | 5 31 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 12 35 1 0 0 0 0 | + | 12 35 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 32 33 34 | + | M SAL 1 3 32 33 34 |
| − | M SBL 1 1 38 | + | M SBL 1 1 38 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 38 -0.9209 0.1232 | + | M SBV 1 38 -0.9209 0.1232 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 35 36 | + | M SAL 2 2 35 36 |
| − | M SBL 2 1 40 | + | M SBL 2 1 40 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 40 -0.6706 0.3872 | + | M SBV 2 40 -0.6706 0.3872 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FEAGS0025 | + | ID FL5FEAGS0025 |
| − | FORMULA C23H20O13 | + | FORMULA C23H20O13 |
| − | EXACTMASS 504.090390726 | + | EXACTMASS 504.090390726 |
| − | AVERAGEMASS 504.3971 | + | AVERAGEMASS 504.3971 |
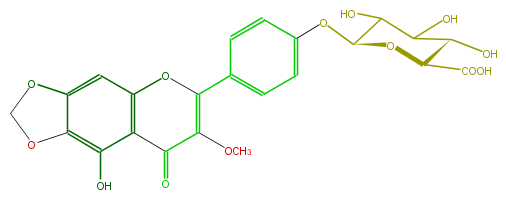
| − | SMILES O=C(C=3OC)c(c(O)1)c(OC(c(c4)ccc(OC(O5)C(O)C(C(O)C5C(O)=O)O)c4)3)cc(O2)c(OC2)1 | + | SMILES O=C(C=3OC)c(c(O)1)c(OC(c(c4)ccc(OC(O5)C(O)C(C(O)C5C(O)=O)O)c4)3)cc(O2)c(OC2)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 40 0 0 0 0 0 0 0 0999 V2000
0.0784 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6361 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6361 1.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0784 1.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7929 1.3320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7929 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3504 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0649 0.5071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7794 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7794 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0649 -1.1429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3504 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4939 0.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2083 0.0946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2083 -0.7304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4939 -1.1429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0649 -1.8499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4939 -1.8834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9928 0.3495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4778 -0.3179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9928 -0.9854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6591 1.7635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0538 1.2026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8789 1.1849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5797 0.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1852 1.3100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3600 1.3278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0903 1.7725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9803 0.9992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0228 1.8834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3963 1.6804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5006 0.6259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4778 0.8376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1952 -0.3263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6799 -1.1176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6799 -1.8520 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
11 17 2 0 0 0 0
16 18 1 0 0 0 0
14 19 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
26 29 1 0 0 0 0
22 30 1 0 0 0 0
5 31 1 0 0 0 0
23 31 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
25 32 1 0 0 0 0
35 36 1 0 0 0 0
12 35 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 32 33 34
M SBL 1 1 38
M SMT 1 COOH
M SBV 1 38 -0.9209 0.1232
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 35 36
M SBL 2 1 40
M SMT 2 OCH3
M SBV 2 40 -0.6706 0.3872
S SKP 5
ID FL5FEAGS0025
FORMULA C23H20O13
EXACTMASS 504.090390726
AVERAGEMASS 504.3971
SMILES O=C(C=3OC)c(c(O)1)c(OC(c(c4)ccc(OC(O5)C(O)C(C(O)C5C(O)=O)O)c4)3)cc(O2)c(OC2)1
M END