Mol:FL5FDGGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0287 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0287 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0287 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0287 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3143 -1.8267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3143 -1.8267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5998 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5998 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5998 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5998 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3143 -0.1768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3143 -0.1768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8854 -1.8267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8854 -1.8267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1709 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1709 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1709 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1709 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8854 -0.1768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8854 -0.1768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8840 -2.6363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8840 -2.6363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5433 -0.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5433 -0.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2715 -0.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2715 -0.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9997 -0.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9997 -0.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9997 0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9997 0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2715 1.0844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2715 1.0844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5433 0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5433 0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3143 -2.6514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3143 -2.6514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8192 1.1371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8192 1.1371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6676 -0.1706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6676 -0.1706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7276 -0.5971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7276 -0.5971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2715 1.9250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2715 1.9250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8645 -0.2681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8645 -0.2681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4851 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4851 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2148 -0.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2148 -0.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9486 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9486 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3281 -0.2681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3281 -0.2681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5985 -0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5985 -0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2041 -0.2854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2041 -0.2854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0854 0.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0854 0.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9800 -0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9800 -0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2998 0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2998 0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4930 -0.3232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4930 -0.3232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7490 0.5726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7490 0.5726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4930 1.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4930 1.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2998 1.9399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2998 1.9399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0438 1.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0438 1.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9610 2.6514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9610 2.6514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5244 2.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5244 2.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7270 1.6496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7270 1.6496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9800 0.1443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9800 0.1443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6188 -1.8702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6188 -1.8702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7310 -2.5123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7310 -2.5123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 14 21 1 0 0 0 0 | + | 14 21 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 21 24 1 0 0 0 0 | + | 21 24 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 8 42 1 0 0 0 0 | + | 8 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -0.7897 0.4560 | + | M SBV 1 47 -0.7897 0.4560 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FDGGS0004 | + | ID FL5FDGGS0004 |
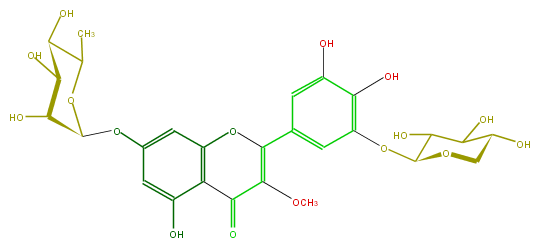
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(c(OC(C5O)OCC(C5O)O)1)c(C(O4)=C(C(=O)c(c43)c(O)cc(c3)OC(C(O)2)OC(C)C(O)C2O)OC)cc(O)c(O)1 | + | SMILES c(c(OC(C5O)OCC(C5O)O)1)c(C(O4)=C(C(=O)c(c43)c(O)cc(c3)OC(C(O)2)OC(C)C(O)C2O)OC)cc(O)c(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-3.0287 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0287 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3143 -1.8267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5998 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5998 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3143 -0.1768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8854 -1.8267 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1709 -1.4142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1709 -0.5892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8854 -0.1768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8840 -2.6363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5433 -0.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2715 -0.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9997 -0.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9997 0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2715 1.0844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5433 0.6639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3143 -2.6514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8192 1.1371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6676 -0.1706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7276 -0.5971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2715 1.9250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8645 -0.2681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4851 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2148 -0.7167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9486 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3281 -0.2681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5985 -0.4766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2041 -0.2854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0854 0.0999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9800 -0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2998 0.1427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4930 -0.3232 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7490 0.5726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4930 1.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2998 1.9399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0438 1.0440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9610 2.6514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5244 2.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7270 1.6496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9800 0.1443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6188 -1.8702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7310 -2.5123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
1 20 1 0 0 0 0
14 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
21 24 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
37 40 1 0 0 0 0
32 41 1 0 0 0 0
33 20 1 0 0 0 0
42 43 1 0 0 0 0
8 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -0.7897 0.4560
S SKP 5
ID FL5FDGGS0004
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(c(OC(C5O)OCC(C5O)O)1)c(C(O4)=C(C(=O)c(c43)c(O)cc(c3)OC(C(O)2)OC(C)C(O)C2O)OC)cc(O)c(O)1
M END