Mol:FL5FCDGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3806 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3806 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3806 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3806 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8243 -1.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8243 -1.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2680 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2680 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2680 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2680 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8243 0.1397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8243 0.1397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7117 -1.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7117 -1.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1554 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1554 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1554 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1554 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7117 0.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7117 0.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7117 -1.6459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7117 -1.6459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5993 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5993 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0323 -0.1878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0323 -0.1878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5347 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5347 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5347 0.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5347 0.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0323 1.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0323 1.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5993 0.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5993 0.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8243 -1.7872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8243 -1.7872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5993 -1.1449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5993 -1.1449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5347 0.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5347 0.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8362 1.5443 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.8362 1.5443 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.5408 1.0327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.5408 1.0327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1089 1.1951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1089 1.1951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6803 1.0327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6803 1.0327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9758 1.5443 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9758 1.5443 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4077 1.3821 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4077 1.3821 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6421 2.0779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6421 2.0779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4485 1.8481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4485 1.8481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4020 1.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4020 1.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7378 0.4373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7378 0.4373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2377 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2377 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2511 1.6977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2511 1.6977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6925 2.5950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6925 2.5950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8750 0.7489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8750 0.7489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8749 -0.0760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8749 -0.0760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 16 32 1 0 0 0 0 | + | 16 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 24 34 1 0 0 0 0 | + | 24 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 34 35 | + | M SAL 3 2 34 35 |
| − | M SBL 3 1 37 | + | M SBL 3 1 37 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 37 3.0234 1.0594 | + | M SVB 3 37 3.0234 1.0594 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 35 | + | M SBL 2 1 35 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 35 0.2511 1.6977 | + | M SVB 2 35 0.2511 1.6977 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 30 31 | + | M SAL 1 2 30 31 |
| − | M SBL 1 1 33 | + | M SBL 1 1 33 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 33 -3.7378 0.4373 | + | M SVB 1 33 -3.7378 0.4373 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FCDGS0001 | + | ID FL5FCDGS0001 |
| − | KNApSAcK_ID C00005608 | + | KNApSAcK_ID C00005608 |
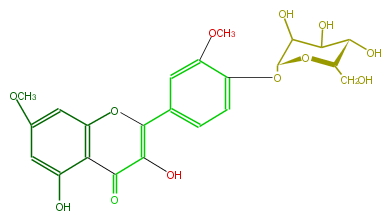
| − | NAME Rhamnazin 4'-glucoside | + | NAME Rhamnazin 4'-glucoside |
| − | CAS_RN 80651-75-8 | + | CAS_RN 80651-75-8 |
| − | FORMULA C23H24O12 | + | FORMULA C23H24O12 |
| − | EXACTMASS 492.126776232 | + | EXACTMASS 492.126776232 |
| − | AVERAGEMASS 492.42946 | + | AVERAGEMASS 492.42946 |
| − | SMILES OC(C4O)[C@@H](O[C@H](CO)[C@@H]4O)Oc(c3)c(OC)cc(c3)C(=C2O)Oc(c1C2=O)cc(cc1O)OC | + | SMILES OC(C4O)[C@@H](O[C@H](CO)[C@@H]4O)Oc(c3)c(OC)cc(c3)C(=C2O)Oc(c1C2=O)cc(cc1O)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-3.3806 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3806 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8243 -1.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2680 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2680 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8243 0.1397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7117 -1.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1554 -0.8239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1554 -0.1815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7117 0.1397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7117 -1.6459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5993 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0323 -0.1878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5347 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5347 0.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0323 1.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5993 0.7943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8243 -1.7872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5993 -1.1449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5347 0.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8362 1.5443 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.5408 1.0327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1089 1.1951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6803 1.0327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9758 1.5443 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4077 1.3821 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6421 2.0779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4485 1.8481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4020 1.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7378 0.4373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2377 1.3034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2511 1.6977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6925 2.5950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8750 0.7489 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8749 -0.0760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
8 19 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
20 15 1 0 0 0 0
1 30 1 0 0 0 0
30 31 1 0 0 0 0
16 32 1 0 0 0 0
32 33 1 0 0 0 0
24 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 34 35
M SBL 3 1 37
M SMT 3 CH2OH
M SVB 3 37 3.0234 1.0594
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 35
M SMT 2 OCH3
M SVB 2 35 0.2511 1.6977
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 30 31
M SBL 1 1 33
M SMT 1 OCH3
M SVB 1 33 -3.7378 0.4373
S SKP 8
ID FL5FCDGS0001
KNApSAcK_ID C00005608
NAME Rhamnazin 4'-glucoside
CAS_RN 80651-75-8
FORMULA C23H24O12
EXACTMASS 492.126776232
AVERAGEMASS 492.42946
SMILES OC(C4O)[C@@H](O[C@H](CO)[C@@H]4O)Oc(c3)c(OC)cc(c3)C(=C2O)Oc(c1C2=O)cc(cc1O)OC
M END